

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: Cn1sc(=O)c2cc(ccc12)S(N)(=O)=O
InChI Key: InChIKey=DFPYCCVFXMWMJM-UHFFFAOYSA-N
PDB links: 1 PDB ID matches this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbonic anhydrase 2 (Homo sapiens (Human)) | BDBM13072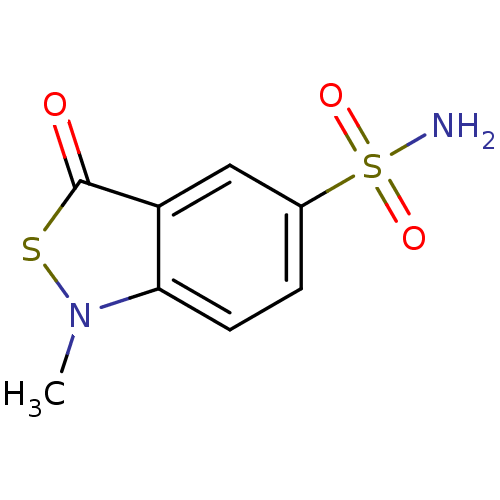 (0134-36 | 1-methyl-3-oxo-1,3-dihydro-2,1-benzothia...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 246 | n/a | n/a | n/a | n/a | 7.4 | 25 |
University of Marburg | Assay Description Initial rates of 4-nitrophenyl acetate hydrolysis catalyzed by different CA isozymes were monitored spectrophotometrically at 400 nm. A molar absorpt... | J Med Chem 45: 3588-602 (2002) Article DOI: 10.1021/jm011112j BindingDB Entry DOI: 10.7270/Q2RJ4GQP | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||