

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM15337 (5R)-6-(4-{[2-(3-iodobenzyl)-3-oxocyclohex-1-en-1-yl]amino}phenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one::(5R)-6-[4-({2-[(3-iodophenyl)methyl]-3-oxocyclohex-1-en-1-yl}amino)phenyl]-5-methyl-2,3,4,5-tetrahydropyridazin-3-one::MERCK1::dihydropyridazinone 14e
SMILES: C[C@@H]1CC(=O)NN=C1c1ccc(NC2=C(Cc3cccc(I)c3)C(=O)CCC2)cc1
InChI Key: InChIKey=QNURTFDBHAQRSI-OAHLLOKOSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phosphodiesterase Type 3 (PDE3B) (Homo sapiens (Human)) | BDBM15337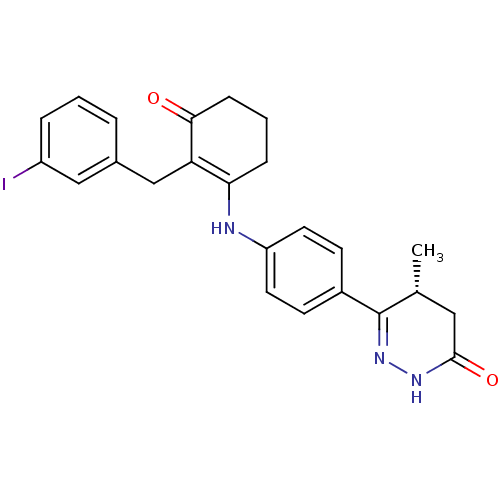 ((5R)-6-(4-{[2-(3-iodobenzyl)-3-oxocyclohex-1-en-1-...) | PDB MMDB KEGG B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 0.110 | n/a | n/a | n/a | n/a | 7.5 | 22 |
Merck Research Laboratories | Assay Description PDE activity was monitored by measuring the hydrolysis of [3H]-cAMP to [3H]-AMP using scintillation proximity assay (SPA). [3H]-AMP was captured by t... | Bioorg Med Chem Lett 13: 3983-7 (2003) Article DOI: 10.1016/j.bmcl.2003.08.056 BindingDB Entry DOI: 10.7270/Q2JD4V11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Phosphodiesterase Type 3 (PDE3B) (Homo sapiens (Human)) | BDBM15337 ((5R)-6-(4-{[2-(3-iodobenzyl)-3-oxocyclohex-1-en-1-...) | PDB MMDB KEGG B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 0.270 | n/a | n/a | n/a | n/a | 7.5 | 22 |
Merck Research Laboratories | Assay Description PDE activity was monitored by measuring the hydrolysis of [3H]-cAMP to [3H]-AMP using scintillation proximity assay (SPA). [3H]-AMP was captured by t... | Biochemistry 43: 6091-100 (2004) Article DOI: 10.1021/bi049868i BindingDB Entry DOI: 10.7270/Q2DR2SQM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Phosphodiesterase Type 3 (PDE3A) (Homo sapiens (Human)) | BDBM15337 ((5R)-6-(4-{[2-(3-iodobenzyl)-3-oxocyclohex-1-en-1-...) | PDB KEGG DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 0.330 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories | Assay Description PDE activity was monitored by measuring the hydrolysis of [3H]-cAMP to [3H]-AMP using scintillation proximity assay (SPA). [3H]-AMP was captured by t... | Bioorg Med Chem Lett 13: 3983-7 (2003) Article DOI: 10.1016/j.bmcl.2003.08.056 BindingDB Entry DOI: 10.7270/Q2JD4V11 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||