

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM191602 3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)isonicotinic acid (N12)::US10040779, Example 52::US10336727, Example 52::US9617242, Example 52
SMILES: Cn1cc(Nc2cnccc2C(O)=O)c2cccnc12
InChI Key: InChIKey=RBGNHCYGFBRIRO-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lysine-specific demethylase 5A (KDM5A(aa 1-739)ΔAP(2C-2S)) (Homo sapiens (Human)) | BDBM191602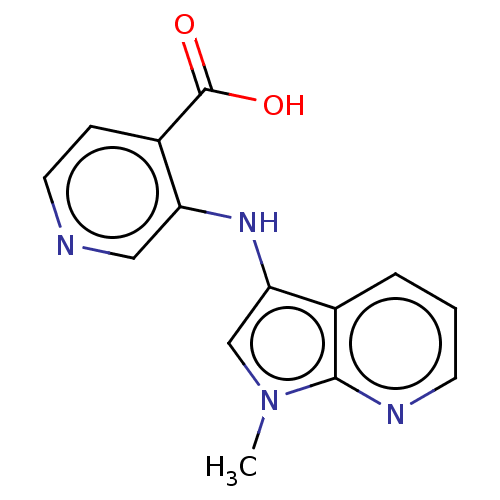 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 6.00E+3 | n/a | n/a | n/a | n/a | n/a | 25 |
Emory University | Assay Description For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 4C (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | US Patent | n/a | n/a | 550 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Quanticel Research Inc US Patent | Assay Description The ability of test compounds to inhibit the activity of JMJD2C was determined in 384-well plate format under the following reaction conditions: 0.3 ... | US Patent US10336727 (2019) BindingDB Entry DOI: 10.7270/Q2571FBT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5C (KDM5C(aa 1-789)ΔAP) (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | 1.90E+4 | n/a | n/a | n/a | n/a | n/a | 25 |
Emory University | Assay Description For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5D (KDM5D(aa 1-760)ΔAP) (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | 2.20E+4 | n/a | n/a | n/a | n/a | n/a | 25 |
Emory University | Assay Description For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5A (KDM5A(aa 1-588)ΔAP) (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 450 | n/a | n/a | n/a | n/a | n/a |
Emory University | Assay Description ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 5B (KDM5B(aa 1-604)ΔAP) (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 330 | n/a | n/a | n/a | n/a | n/a |
Emory University | Assay Description ITC experiments were carried out at an enzyme [E] concentration of 25-50 μM with 0.25-1 mM compound [I] concentration in the KDM5 storage buffer... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 5A (KDM5A(aa 1-1090)-Flag) (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 700 | n/a | n/a | n/a | n/a | 7.2 | 25 |
Emory University | Assay Description Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 5B (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | 780 | n/a | n/a | n/a | n/a | 7.2 | 25 |
Emory University | Assay Description Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5C (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | 1.40E+3 | n/a | n/a | n/a | n/a | 7.2 | 25 |
Emory University | Assay Description Reactions were performed in 50 mM HEPES [pH 7.2 for KDM5 or pH 7.5 for KDM4A(1-350)] containing 0.01% BSA and 0.01% Tween-20. To each well, 3 μL... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5A (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB US Patent | n/a | n/a | <0.100 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Quanticel Research, Inc. US Patent | Assay Description The enzymatic assay of Jarid1A activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test c... | US Patent US10040779 (2018) BindingDB Entry DOI: 10.7270/Q26H4KF1 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 5B (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | US Patent | n/a | n/a | <0.100 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Quanticel Research, Inc. US Patent | Assay Description The ability of test compounds to inhibit the activity of Jarid1B was determined in 384-well plate format under the following reaction conditions: 0.8... | US Patent US10040779 (2018) BindingDB Entry DOI: 10.7270/Q26H4KF1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 4C (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | US Patent | n/a | n/a | 0.550 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Quanticel Research, Inc. US Patent | Assay Description The ability of test compounds to inhibit the activity of JMJD2C was determined in 384-well plate format under the following reaction conditions: 0.3 ... | US Patent US10040779 (2018) BindingDB Entry DOI: 10.7270/Q26H4KF1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5A (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB US Patent | n/a | n/a | <100 | n/a | n/a | n/a | n/a | n/a | n/a |
CELGENE QUANTICEL RESEARCH, INC. US Patent | Assay Description The enzymatic assay of Jarid1A activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test c... | US Patent US9617242 (2017) BindingDB Entry DOI: 10.7270/Q2XK8HM5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 5B (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | US Patent | n/a | n/a | <100 | n/a | n/a | n/a | n/a | n/a | n/a |
CELGENE QUANTICEL RESEARCH, INC. US Patent | Assay Description The ability of test compounds to inhibit the activity of Jarid1B was determined in 384-well plate format under the following reaction conditions: 0.8... | US Patent US9617242 (2017) BindingDB Entry DOI: 10.7270/Q2XK8HM5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 4C (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | US Patent | n/a | n/a | 550 | n/a | n/a | n/a | n/a | n/a | n/a |
CELGENE QUANTICEL RESEARCH, INC. US Patent | Assay Description The ability of test compounds to inhibit the activity of JMJD2C was determined in 384-well plate format under the following reaction conditions: 0.3 ... | US Patent US9617242 (2017) BindingDB Entry DOI: 10.7270/Q2XK8HM5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5A (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB US Patent | n/a | n/a | <100 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Quanticel Research Inc US Patent | Assay Description The enzymatic assay of Jarid1A activity is based upon Time Resolved-Fluorescence Resonance Energy Transfer (TR-FRET) detection. The ability of test c... | US Patent US10336727 (2019) BindingDB Entry DOI: 10.7270/Q2571FBT | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Lysine-specific demethylase 5B (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | US Patent | n/a | n/a | <100 | n/a | n/a | n/a | n/a | n/a | n/a |
Celgene Quanticel Research Inc US Patent | Assay Description The ability of test compounds to inhibit the activity of Jarid1B was determined in 384-well plate format under the following reaction conditions: 0.8... | US Patent US10336727 (2019) BindingDB Entry DOI: 10.7270/Q2571FBT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Lysine-specific demethylase 5B (KDM5B(aa 1-755)ΔAP) (Homo sapiens (Human)) | BDBM191602 (3-((1-methyl-1Hpyrrolo[2,3-b]pyridin-3-yl)amino)is...) | PDB GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | 2.30E+3 | n/a | n/a | n/a | n/a | n/a | 25 |
Emory University | Assay Description For inhibition assays, powders of inhibitor compounds were dissolved in 100% (v/v) DMSO to give a 20 or 50 mM concentration and stored at -20 °C unti... | Cell Chem Biol 23: 769-81 (2016) Article DOI: 10.1016/j.chembiol.2016.06.006 BindingDB Entry DOI: 10.7270/Q2BZ64VH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||