

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM20714 (2S)-N-[(2S,3S,4S,5S)-6-cyclohexyl-3,4-dihydroxy-5-[(2S)-3-methyl-2-{[methyl(pyridin-2-ylmethyl)carbamoyl]amino}butanamido]-1-phenylhexan-2-yl]-3-methyl-2-{[methyl(pyridin-2-ylmethyl)carbamoyl]amino}butanamide::Inhibitor PC
SMILES: CC(C)[C@H](NC(=O)N(C)Cc1ccccn1)C(=O)N[C@@H](CC1CCCCC1)[C@H](O)[C@@H](O)[C@H](Cc1ccccc1)NC(=O)[C@@H](NC(=O)N(C)Cc1ccccn1)C(C)C
InChI Key: InChIKey=JQIFSYRTTKZQMY-UNHORJANSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 Protease (Human immunodeficiency virus type 1) | BDBM20714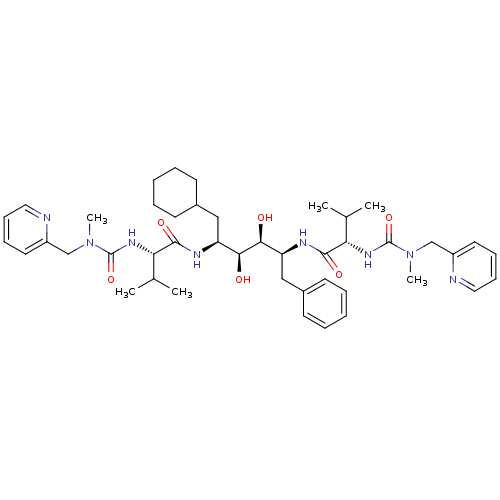 ((2S)-N-[(2S,3S,4S,5S)-6-cyclohexyl-3,4-dihydroxy-5...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 3.20 | -12.0 | n/a | n/a | n/a | n/a | n/a | 4.7 | 37 |
Universidad de Santiago de Compostela | Assay Description The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe... | J Med Chem 51: 852-60 (2008) Article DOI: 10.1021/jm701170f BindingDB Entry DOI: 10.7270/Q2QN652X | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 Protease Mutant (V82A) (Human immunodeficiency virus type 1) | BDBM20714 ((2S)-N-[(2S,3S,4S,5S)-6-cyclohexyl-3,4-dihydroxy-5...) | PDB MMDB B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 14 | -11.1 | n/a | n/a | n/a | n/a | n/a | 4.7 | 37 |
Universidad de Santiago de Compostela | Assay Description The inhibition constants Ki were determined by monitoring the inhibition of hydrolysis of the chromogenic substrate using a Hewlett-Packard 8452A spe... | J Med Chem 51: 852-60 (2008) Article DOI: 10.1021/jm701170f BindingDB Entry DOI: 10.7270/Q2QN652X | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||