

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: OC(=O)c1ccc(OC[C@H](C2CCCCC2)n2c(nc3cc(F)c(F)cc23)-c2ccc(Cl)cc2)c(F)c1
InChI Key: InChIKey=CPOSGAZIIDJVMJ-RUZDIDTESA-N
PDB links: 1 PDB ID matches this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bile acid receptor (Homo sapiens (Human)) | BDBM225938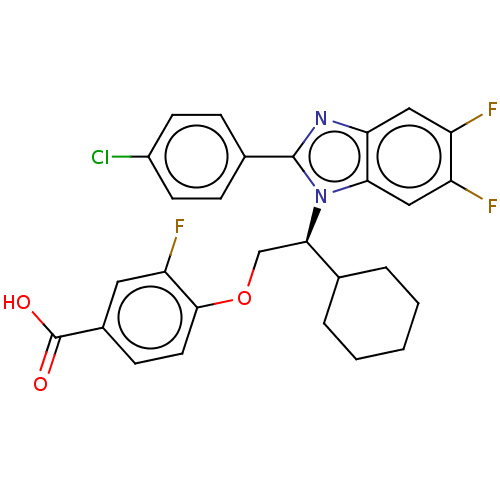 (FXR_28) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB D3R | n/a | n/a | 21 | n/a | n/a | n/a | n/a | 7.4 | n/a |
D3R | Assay Description The assay buffer contained 50 mM HEPES (pH 7.4), 10 mM NaCl, 5 mM MgCl2 and 0.01% CHAPS. The reactions were incubated for 30 min in the presence of [... | D3R 882: (2017) BindingDB Entry DOI: 10.7270/Q2MC8XWV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||