

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: C[NH2+]CCCC[C@H](NC(=O)[C@@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=O)[C@@H](N)CCSC)[C@@H](C)O)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H](C)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CO)C(=O)N[C@@H]([C@@H](C)O)C(=O)NCC(=O)NCC(=O)N[C@@H](CCCCN)C(=O)N[C@@H](C)C(=O)N1CCC[C@H]1C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CCC(N)=O)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](C)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](C)C(O)=O
InChI Key: InChIKey=FLVCSJXGQHMODF-MLFSIEHMSA-O
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histone-lysine N-methyltransferase PRDM9 [195-385] (Homo sapiens (Human)) | BDBM228647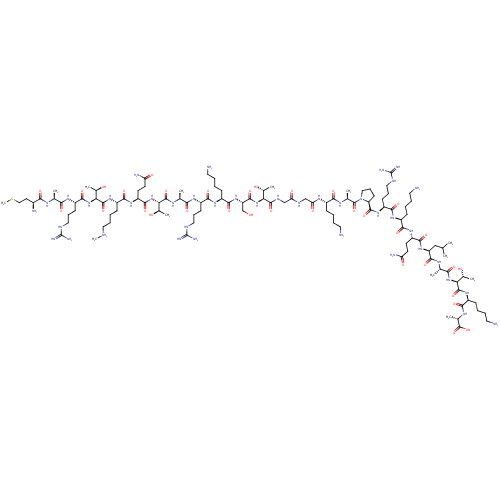 (H3 (1-25) K4me1 | MARTKQTARKSTGGKAPRKQLATKA) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 4.70E+3 | n/a | n/a | n/a | 8.5 | 23 |
University of Toronto | Assay Description In this assay [3H]AdoMet (PerkinElmer Life Sciences; catalog number NET155V250UC)was used as a methyl donor to methylate biotinylated histone peptide... | J Biol Chem 289: 12177-88 (2014) Article DOI: 10.1074/jbc.M113.523183 BindingDB Entry DOI: 10.7270/Q2SQ8Z86 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||