

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: OC(=O)C(CSC(c1ccccc1)(c1ccccc1)c1ccccc1)NC(c1ccccc1)c1ccccc1
InChI Key: InChIKey=YODWYMRZKLKPGM-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinesin-like protein KIF11 (Homo sapiens (Human)) | BDBM23810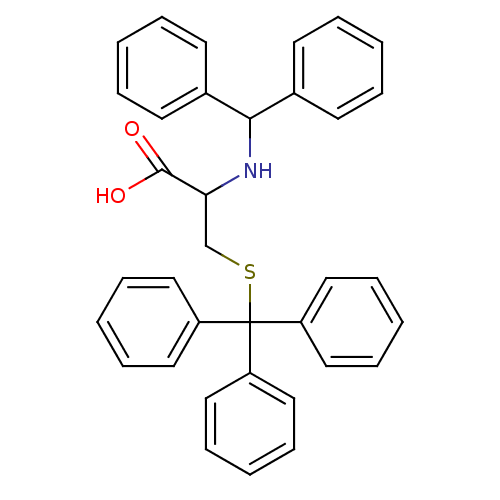 (2-[(diphenylmethyl)amino]-3-[(triphenylmethyl)sulf...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 25 |
Institut de Biologie Structurale | Assay Description The microtubule-activated ATPase rates were measured using the pyruvate kinase/lactate dehydrogenase-linked assay. To optimize the signal for basal E... | J Med Chem 51: 1115-25 (2008) Article DOI: 10.1021/jm070606z BindingDB Entry DOI: 10.7270/Q2DJ5CXD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||