

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM31665 imidazole-dioxolane, 17
SMILES: Clc1ccc(CC[C@@]2(Cn3ccnc3)OC[C@@H](CSc3ccc4ccccc4c3)O2)cc1
InChI Key: InChIKey=PGMLVTZUCFVHFP-AZGAKELHSA-N
Data: 4 IC50
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heme Oxygenase 1 (HO-1) (Rattus norvegicus (rat)) | BDBM31665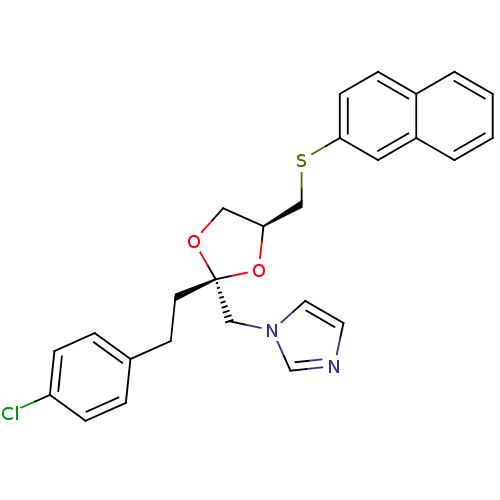 (imidazole-dioxolane, 17) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 900 | n/a | n/a | n/a | n/a | 7.4 | 37 |
Queen's University | Assay Description HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31665 (imidazole-dioxolane, 17) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 2E1 (Rattus norvegicus (rat)) | BDBM31665 (imidazole-dioxolane, 17) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | >1.00E+5 | n/a | n/a | n/a | n/a | 7.4 | n/a |
Queen's University | Assay Description CYP2E1 hydroxylation of p-nitrophenol was determined by the spectrophotometric measurement of 4-nitrocatechol. Briefly, reaction mixture consisting o... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Heme Oxygenase 2 (HO-2) (Rattus norvegicus (rat)) | BDBM31665 (imidazole-dioxolane, 17) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.00E+4 | n/a | n/a | n/a | n/a | 7.4 | n/a |
Queen's University | Assay Description HO activity in rat spleen (HO-1) and brain (HO-2) microsomal fractions was determined by the quantitation of CO formed from the degradation of methem... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||