

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM32652 m-chlorobenzylamide, 1
SMILES: CC[C@@H](N)C(=O)N1CCCC1C(=O)NCc1cccc(Cl)c1
InChI Key: InChIKey=OTYYXGRJXJFTCD-KWCCSABGSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prothrombin (Homo sapiens (Human)) | BDBM32652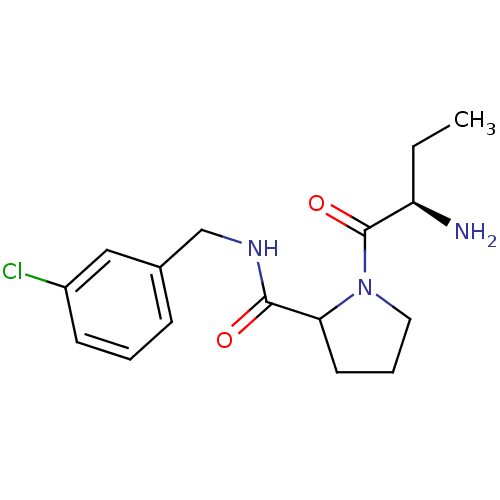 (m-chlorobenzylamide, 1) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 6.80E+3 | -7.04 | n/a | n/a | n/a | n/a | n/a | 7.4 | 25 |
Philipps-University Marburg | Assay Description Kinetic inhibition of human thrombin was determined photometrically at 405 nm using the chromogenic substrate Pefachrom tPa. Reactions were performed... | J Mol Biol 391: 552-64 (2009) Article DOI: 10.1016/j.jmb.2009.06.016 BindingDB Entry DOI: 10.7270/Q2RV0M1Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Cell (A) | Syringe (B) | Cell Links | Syringe Links | Cell + Syr Links | ΔG° kcal/mole | -TΔS° kcal/mole | ΔH° kcal/mole | log K | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|
| Thrombin (Homo sapiens (Human)) | BDBM32652 (m-chlorobenzylamide, 1) | GoogleScholar PDB | PC cid PC sid PDB | -7.48 | 0.527 | -8.00 | n/a | 7.80 | 25 | |
Philipps-University Marburg | J Mol Biol 391: 552-64 (2009) | |||||||||