

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: CCNC(=O)Nc1nc2cc(c(N[C@H]3CCOC3)nc2s1)-c1ccn(CCOC)c(=O)c1
InChI Key: InChIKey=BQWMHHFWBYWCQI-AWEZNQCLSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA gyrase subunit B (Mycobacterium smegmatis) | BDBM50010338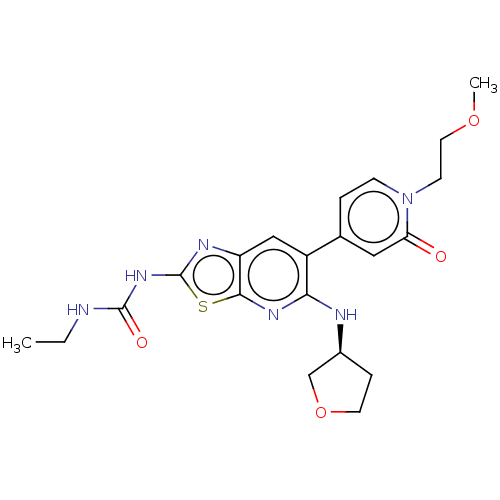 (CHEMBL3259858) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 3.70 | n/a | n/a | n/a | n/a | n/a | n/a |
AstraZeneca India Pvt. Ltd. Curated by ChEMBL | Assay Description Inhibition of Mycobacterium smegmatis DNA gyrase B ATPase activity assessed as inorganic phosphate release using ATP as substrate by colorimetric ana... | Cell Chem Biol 56: 8834-48 (2013) Article DOI: 10.1021/jm401268f BindingDB Entry DOI: 10.7270/Q2RV0Q7W | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||