

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: C#CCn1ncc(n1)C1CCCNC1
InChI Key: InChIKey=FRGSXTHALSHQFX-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50038217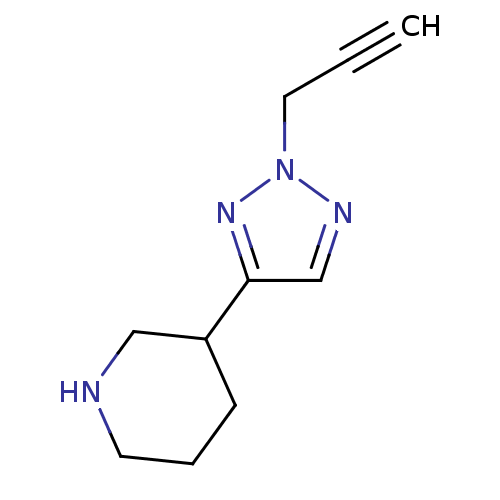 (3-(2-Prop-2-ynyl-2H-[1,2,3]triazol-4-yl)-piperidin...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 84 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
H. Lundbeck A/S Curated by ChEMBL | Assay Description In vitro binding affinity for muscarinic M1 receptor by displacing [3H]-Pirenzepine binding on rat brain homogenate. | J Med Chem 37: 4085-99 (1995) BindingDB Entry DOI: 10.7270/Q21N8066 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50038217 (3-(2-Prop-2-ynyl-2H-[1,2,3]triazol-4-yl)-piperidin...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 140 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
H. Lundbeck A/S Curated by ChEMBL | Assay Description In vitro binding affinity for muscarinic M1 receptor by displacing [3H]-Pirenzepine binding on rat brain homogenate. | J Med Chem 37: 4085-99 (1995) BindingDB Entry DOI: 10.7270/Q21N8066 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||