

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: Nc1ncnc2n(cnc12)[C@@H]1O[C@H](COP(O)(=O)O[C@@H]2[C@H](O)[C@@H](COP(O)(=O)O[C@@H]3[C@H](O)[C@@H](COP(O)(O)=O)O[C@H]3n3cnc4c(N)ncnc34)O[C@H]2n2cnc3c(N)ncnc23)[C@@H](O)[C@H]1O
InChI Key: InChIKey=SIIZPVYVXNXXQG-UQTMIEBXSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2-5A-dependent ribonuclease (Homo sapiens (Human)) | BDBM50085554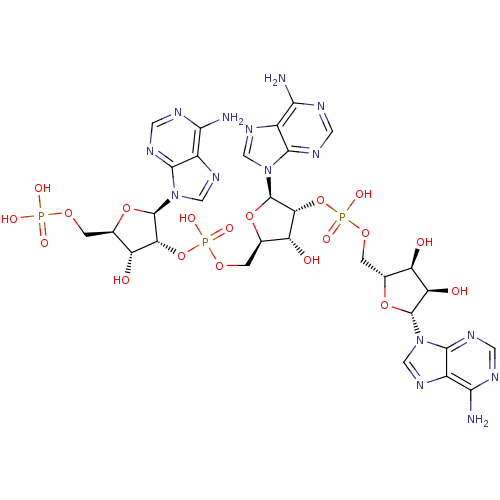 (5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5'...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid UniChem Similars | MMDB PubMed | n/a | n/a | n/a | n/a | 0.200 | n/a | n/a | n/a | n/a |
Gifu University Curated by ChEMBL | Assay Description Activation of purified recombinant human Ribonuclease L by the compound was measured as degradation of [32P]-pC11U2C7 | Bioorg Med Chem Lett 10: 329-31 (2000) BindingDB Entry DOI: 10.7270/Q2TM79B4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 2-5A-dependent ribonuclease (Homo sapiens (Human)) | BDBM50085554 (5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5'...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid UniChem Similars | MMDB PubMed | n/a | n/a | 7.30 | n/a | n/a | n/a | n/a | n/a | n/a |
Gifu University Curated by ChEMBL | Assay Description Activation of purified recombinant human Ribonuclease L by the compound was measured as degradation of [32P]-pC11U2C7 | Bioorg Med Chem Lett 10: 329-31 (2000) BindingDB Entry DOI: 10.7270/Q2TM79B4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 2-5A-dependent ribonuclease (Homo sapiens (Human)) | BDBM50085554 (5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5'...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid UniChem Similars | MMDB PubMed | n/a | n/a | n/a | n/a | 0.150 | n/a | n/a | n/a | n/a |
Gifu University Curated by ChEMBL | Assay Description Activation of purified recombinant human Ribonuclease L by the compound was measured as degradation of poly (U) 3'[32P]p5'C3'p | Bioorg Med Chem Lett 10: 329-31 (2000) BindingDB Entry DOI: 10.7270/Q2TM79B4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||