

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50330317 5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S)-6-{8-chloro-4-hydroxy-3-[4-hydroxy-3-(3-methyl-but-2-enyl)-benzoylamino]-2-oxo-2H-chromen-7-yloxy}-5-hydroxy-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester::5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S)-6-{8-chloro-4-hydroxy-3-[4-hydroxy-3-(4-methyl-pent-3-enyl)-benzoylamino]-2-oxo-2H-chromen-7-yloxy}-5-hydroxy-3-methoxy-2,2-dimethyl-tetrahydro-pyran-4-yl ester::CHEMBL303984::Clorobiocin
SMILES: CO[C@@H]1[C@@H](OC(=O)c2ccc(C)[nH]2)[C@@H](O)[C@H](Oc2ccc3c(O)c(NC(=O)c4ccc(O)c(CC=C(C)C)c4)c(=O)oc3c2Cl)OC1(C)C
InChI Key: InChIKey=FJAQNRBDVKIIKK-LFLQOBSNSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA Gyrase Subunit B (Escherichia coli (strain K12)) | BDBM50330317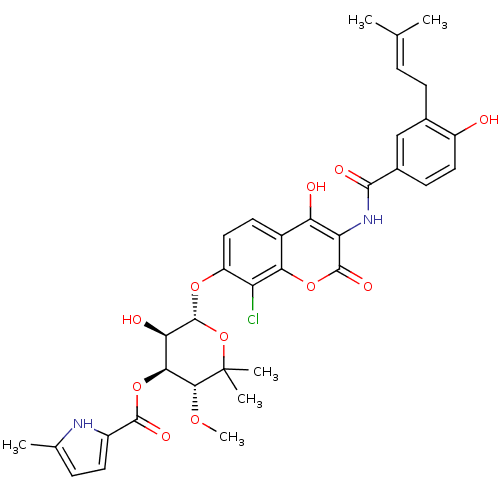 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 73 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA Gyrase (Staphylococcus aureus) | BDBM50330317 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 8.5 | n/a | n/a | n/a | n/a | n/a | n/a |
Bharathiar University Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase assessed as reduction in relaxation of pBR322 DNA by SDS-PAGE analysis | Bioorg Med Chem 26: 3438-3452 (2018) Article DOI: 10.1016/j.bmc.2018.05.016 BindingDB Entry DOI: 10.7270/Q2125W5H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA Gyrase Subunit A (Staphylococcus aureus) | BDBM50330317 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 929 | n/a | n/a | n/a | n/a | n/a | n/a |
VIT University Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase A expressed in Escherichia coli BL21(DE3) using relaxed pBR322 DNA as substrate incubated for 1 hr by ... | Bioorg Med Chem 25: 1448-1455 (2017) Article DOI: 10.1016/j.bmc.2017.01.007 BindingDB Entry DOI: 10.7270/Q2RJ4MNF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli) | BDBM50330317 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 1.20 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli GyrB | J Med Chem 54: 915-29 (2011) Article DOI: 10.1021/jm101121s BindingDB Entry DOI: 10.7270/Q2NS0W23 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||