

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50362007 CHEMBL1939715
SMILES: C[N+](C)(CC[C@H](N)C(=O)NCc1ccc(CNC(=O)[C@@H](N)CC[N+](C)(C)C[C@H]2O[C@H]([C@H](O)[C@@H]2O)n2cnc3c(N)ncnc23)cc1)C[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12
InChI Key: InChIKey=CIKHPSDRFNMMRL-LZFDDTNSSA-P
Data: 1 EC50
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Met repressor (Escherichia coli) | BDBM50362007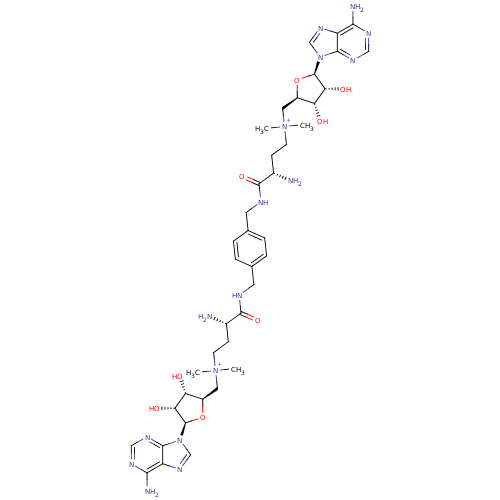 (CHEMBL1939715) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 15 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||