 Found 9 hits for monomerid = 50450592
Found 9 hits for monomerid = 50450592 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kcal/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Muscarinic acetylcholine receptor M2
(RAT) | BDBM50450592
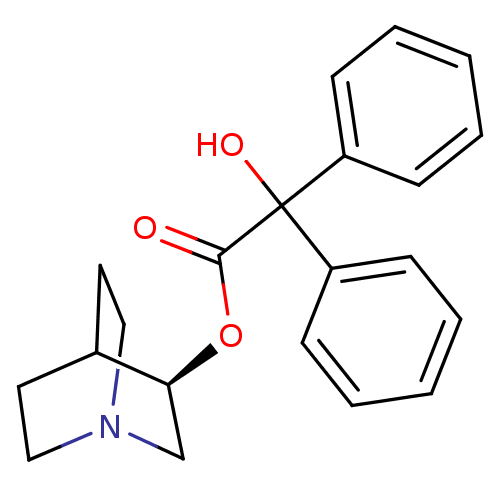
(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Virginia Commonwealth University
Curated by ChEMBL
| Assay Description
Displacement of [3H](-)-quinuclidinyl benzilate(QNB) from muscarinic (M2) receptor in rat heart homogenates |
J Med Chem 34: 2984-9 (1991)
BindingDB Entry DOI: 10.7270/Q27H1K5M |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M2
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory
Curated by ChEMBL
| Assay Description
Ability to displace [3H](-)-quinuclidinyl bezilate(QNB) from M2 receptor in rat heart homogenate |
J Med Chem 36: 848-54 (1993)
BindingDB Entry DOI: 10.7270/Q2S46SM0 |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M2
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| Article
| 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Displacement of [3H]QNB (quinuclidinyl benzylate) from muscarinic M2 receptor of rat heart homogenates. |
Bioorg Med Chem Lett 7: 979-984 (1997)
Article DOI: 10.1016/S0960-894X(97)00143-1
BindingDB Entry DOI: 10.7270/Q2N29XFM |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M1
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | Reactome pathway
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| 0.150 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory
Curated by ChEMBL
| Assay Description
Ability to displace [3H]pirenzepine (PZ) from M1 receptor in rat cortex homogenate |
J Med Chem 36: 848-54 (1993)
BindingDB Entry DOI: 10.7270/Q2S46SM0 |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M1
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | Reactome pathway
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| Article
| 0.150 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Displacement of [3H]pirenzepine from muscarinic M1 receptor of rat cortex homogenates. |
Bioorg Med Chem Lett 7: 979-984 (1997)
Article DOI: 10.1016/S0960-894X(97)00143-1
BindingDB Entry DOI: 10.7270/Q2N29XFM |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M1
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | Reactome pathway
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| 0.150 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Virginia Commonwealth University
Curated by ChEMBL
| Assay Description
Displacement of [3H]pirenzepine from muscarinic acetylcholine receptor M1 in rat cortex homogenates |
J Med Chem 34: 2984-9 (1991)
BindingDB Entry DOI: 10.7270/Q27H1K5M |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M3
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| 0.160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory
Curated by ChEMBL
| Assay Description
Ability to displace [3H]N-methylscopolamine (NMS) from M3 receptor in rat submaxillary gland homogenate |
J Med Chem 36: 848-54 (1993)
BindingDB Entry DOI: 10.7270/Q2S46SM0 |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M2
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | PDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| n/a | n/a | n/a | 0.200 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL)
Curated by ChEMBL
| Assay Description
In vitro binding affinity towards Muscarinic acetylcholine receptor M2 using [3H]-QNB as radioligand from rat heart tissue |
J Med Chem 38: 3908-17 (1995)
BindingDB Entry DOI: 10.7270/Q24M9567 |
More data for this
Ligand-Target Pair | |
Muscarinic acetylcholine receptor M1
(RAT) | BDBM50450592

(CHEMBL558910)Show SMILES OC(C(=O)O[C@H]1CN2CCC1CC2)(c1ccccc1)c1ccccc1 |r,wD:5.4,(8.38,-12.97,;7.3,-14.07,;8.63,-14.83,;8.64,-16.37,;9.97,-14.06,;11.3,-14.82,;11.3,-16.36,;12.63,-17.13,;13.96,-16.36,;13.96,-14.82,;12.63,-14.05,;13.05,-15.29,;12,-15.64,;5.97,-14.84,;4.63,-14.07,;3.3,-14.84,;3.31,-16.38,;4.65,-17.15,;5.98,-16.37,;7.29,-12.53,;8.62,-11.76,;8.62,-10.22,;7.28,-9.45,;5.95,-10.23,;5.96,-11.77,)| Show InChI InChI=1S/C21H23NO3/c23-20(25-19-15-22-13-11-16(19)12-14-22)21(24,17-7-3-1-4-8-17)18-9-5-2-6-10-18/h1-10,16,19,24H,11-15H2/t19-/m0/s1 | Reactome pathway
KEGG
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PubMed
| n/a | n/a | n/a | 0.320 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL)
Curated by ChEMBL
| Assay Description
In vitro binding affinity towards Muscarinic acetylcholine receptor M1 using [3H]QNB as radioligand from rat heart tissue |
J Med Chem 38: 3908-17 (1995)
BindingDB Entry DOI: 10.7270/Q24M9567 |
More data for this
Ligand-Target Pair | |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data