

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM5594 2-arylamino-pyrimidine deriv. 9d::4-{[4-amino-6-(cyclohexylmethoxy)-5-nitrosopyrimidin-2-yl]amino}benzamide::CHEMBL101801
SMILES: NC(=O)c1ccc(Nc2nc(N)c(N=O)c(OCC3CCCCC3)n2)cc1
InChI Key: InChIKey=YBKLJTXDSBEVRV-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyclin-dependent kinase 2 (Homo sapiens (Human)) | BDBM5594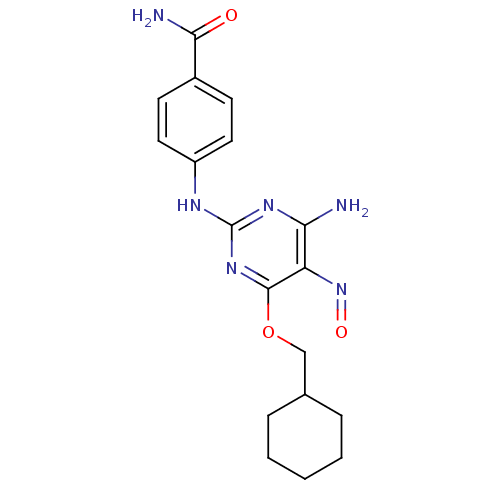 (2-arylamino-pyrimidine deriv. 9d | 4-{[4-amino-6-(...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 2.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Cambridge Curated by ChEMBL | Assay Description Inhibition of CDK2 | J Med Chem 49: 5141-53 (2006) Article DOI: 10.1021/jm060190+ BindingDB Entry DOI: 10.7270/Q27S7NDM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Cyclin-Dependent Kinase 1 (CDK1) (Marthasterias glacialis (starfish)) | BDBM5594 (2-arylamino-pyrimidine deriv. 9d | 4-{[4-amino-6-(...) | UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | n/a | n/a | 70 | n/a | n/a | n/a | n/a | 7.5 | 30 |
University of Newcastle | Assay Description The enzyme was assayed with substrate histone H1 in the presence of 12.5 uM ATP/[gamma-32P] ATP. IC50 is the inhibitor concentration, which inhibits ... | Bioorg Med Chem Lett 13: 3079-82 (2003) Article DOI: 10.1016/s0960-894x(03)00651-6 BindingDB Entry DOI: 10.7270/Q2R78CDT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cyclin-Dependent Kinase 2 (CDK2) (Homo sapiens (Human)) | BDBM5594 (2-arylamino-pyrimidine deriv. 9d | 4-{[4-amino-6-(...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | n/a | n/a | 34 | n/a | n/a | n/a | n/a | 7.5 | 30 |
University of Newcastle | Assay Description The enzyme was assayed with substrate histone H1 in the presence of 12.5 uM ATP/[gamma-32P] ATP. IC50 is the inhibitor concentration, which inhibits ... | Bioorg Med Chem Lett 13: 3079-82 (2003) Article DOI: 10.1016/s0960-894x(03)00651-6 BindingDB Entry DOI: 10.7270/Q2R78CDT | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||