

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM7094 3-Aminopyrazole deriv. 4::N-(5-Cyclopropyl-1H-pyrazol-3-yl)-2-fluoro-2-phenylacetamide
SMILES: FC(C(=O)Nc1cc(n[nH]1)C1CC1)c1ccccc1
InChI Key: InChIKey=QFPGZNFSVUUBNZ-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyclin-Dependent Kinase 2 (CDK2) (Homo sapiens (Human)) | BDBM7094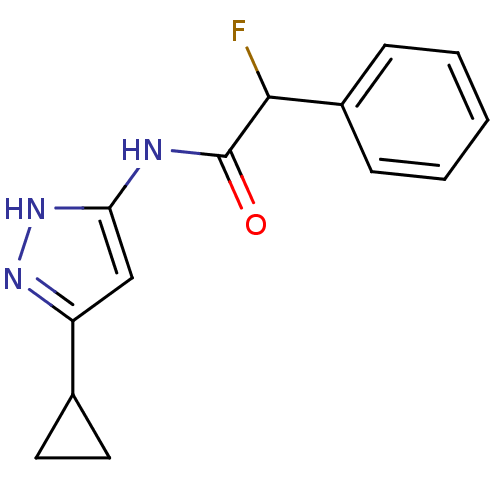 (3-Aminopyrazole deriv. 4 | N-(5-Cyclopropyl-1H-pyr...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt UniProtKB/TrEMBL B.MOAD DrugBank antibodypedia antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 99 | n/a | n/a | n/a | n/a | 7.4 | 22 |
Nerviano Medical Sciences | Assay Description The biochemical activity of compounds was determined by incubation with specific enzymes and substrates in the presence ATP/[gamma-33P] ATP. After in... | J Med Chem 48: 2944-56 (2005) Article DOI: 10.1021/jm0408870 BindingDB Entry DOI: 10.7270/Q2WQ020R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||