

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM853 (2R,3R,4R,5R)-3,4-dihydroxy-N,N'-bis[(1S)-2-methyl-1-(methylcarbamoyl)propyl]-2,5-bis({[4-(1,3-thiazol-2-yl)phenyl]methoxy})hexanediamide::C2-Symmetric inhibitor 11::CHEMBL340116::N1,N6-Bis[(1S)-2-methyl-1-(methylcarbamoyl)propyl]-(2R,3R,4R,5R)-2,5-bis[4-(2-thiazolyl)benzyloxy]-3,4-dihydroxyhexanediamide
SMILES: CNC(=O)[C@@H](NC(=O)[C@H](OCc1ccc(cc1)-c1nccs1)[C@H](O)[C@@H](O)[C@@H](OCc1ccc(cc1)-c1nccs1)C(=O)N[C@@H](C(C)C)C(=O)NC)C(C)C
InChI Key: InChIKey=XISQZIYLHTUXPS-VQKSECAYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human immunodeficiency virus type 1 protease (Human immunodeficiency virus type 1) | BDBM853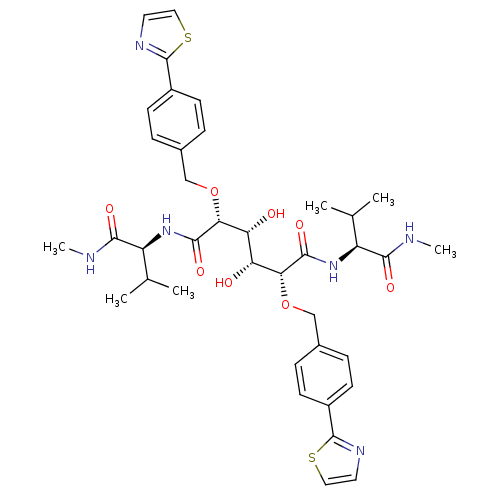 ((2R,3R,4R,5R)-3,4-dihydroxy-N,N'-bis[(1S)-2-methyl...) | PDB MMDB UniProtKB/TrEMBL B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | PubMed | 0.550 | n/a | n/a | 0.643 | n/a | n/a | 4.77E+5 | n/a | n/a |
Uppsala University Curated by ChEMBL | Assay Description Association rate constant for the interaction between inhibitor and HIV-1 protease | J Med Chem 45: 5430-9 (2002) BindingDB Entry DOI: 10.7270/Q2GH9JP4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 Protease (Human immunodeficiency virus type 1) | BDBM853 ((2R,3R,4R,5R)-3,4-dihydroxy-N,N'-bis[(1S)-2-methyl...) | PDB MMDB UniProtKB/TrEMBL B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | 0.600 | -12.8 | n/a | n/a | n/a | n/a | n/a | 5.0 | 30 |
Uppsala University | Assay Description Ki values were determined by using a fluorescent substrate (DABCYL-gamma-Abu-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-EDANS). All incubations were performed a... | J Med Chem 42: 3835-44 (1999) Article DOI: 10.1021/jm9910371 BindingDB Entry DOI: 10.7270/Q2T151VX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Human immunodeficiency virus type 1 protease (Human immunodeficiency virus type 1) | BDBM853 ((2R,3R,4R,5R)-3,4-dihydroxy-N,N'-bis[(1S)-2-methyl...) | PDB MMDB UniProtKB/TrEMBL B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | PubMed | n/a | n/a | n/a | n/a | n/a | 0.000303 | n/a | n/a | n/a |
Uppsala University Curated by ChEMBL | Assay Description Association rate constant for the interaction between inhibitor and HIV-1 protease | J Med Chem 45: 5430-9 (2002) BindingDB Entry DOI: 10.7270/Q2GH9JP4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||