

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM92570 Arylamine compound 4
SMILES: Cc1cccc(c1)-n1cnc2cc(N)ccc12
InChI Key: InChIKey=VFSVFGIODYZMOF-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytochrome P450 130 (G243A) (Mycobacterium tuberculosis) | BDBM92570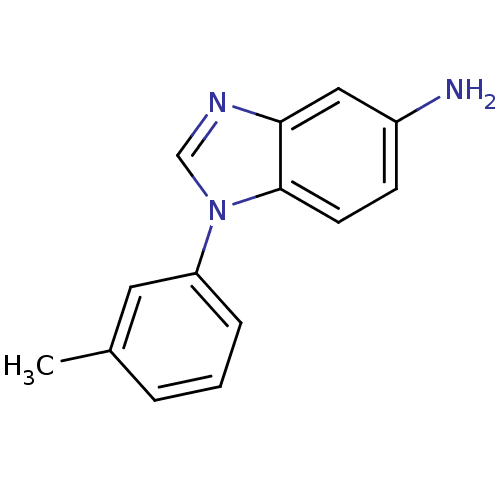 (Arylamine compound 4) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase DrugBank MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 3.26E+4 | n/a | n/a | n/a | n/a | n/a |
University of California, San Francisco | Assay Description Binding assay using CYP130, CYP130 (G234) or CYP142. | J Biol Chem 284: 25211-9 (2009) Article DOI: 10.1074/jbc.M109.017632 BindingDB Entry DOI: 10.7270/Q2RR1WVC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Cytochrome P450 130 (Mycobacterium tuberculosis) | BDBM92570 (Arylamine compound 4) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | n/a | 4.80E+3 | n/a | n/a | n/a | n/a | n/a |
University of California, San Francisco | Assay Description Binding assay using CYP130, CYP130 (G234) or CYP142. | J Biol Chem 284: 25211-9 (2009) Article DOI: 10.1074/jbc.M109.017632 BindingDB Entry DOI: 10.7270/Q2RR1WVC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||