

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM9687 2-methylpropyl N-[(2S)-1-[(1R,2S,5S)-2-({1-[({[(S)-(dimethylcarbamoyl)(phenyl)methyl]carbamoyl}methyl)carbamoyl]-1-oxopentan-2-yl}carbamoyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl]carbamate::SCH446211 (SCH6) Analog 20
SMILES: CCCC(NC(=O)[C@@H]1[C@@H]2[C@H](CN1C(=O)[C@@H](NC(=O)OCC(C)C)C(C)(C)C)C2(C)C)C(=O)C(=O)NCC(=O)N[C@H](C(=O)N(C)C)c1ccccc1
InChI Key: InChIKey=MSHRDJHOMYCZBB-NGNWLLNHSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCV NS3-NS4A Serine Proteinase (Hepatitis C virus (HCV genotype 1a, isolate H)) | BDBM9687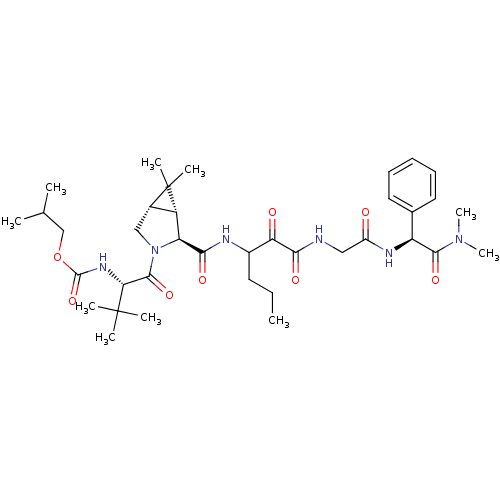 (2-methylpropyl N-[(2S)-1-[(1R,2S,5S)-2-({1-[({[(S)...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 16 | -10.8 | n/a | n/a | n/a | n/a | n/a | 6.5 | 30 |
Schering-Plough Research Institute | Assay Description Proteolytic cleavage of the ester linkage between the Nva (L-norvaline) and the chromophore (PAP) was monitored for change in absorbance at 370 nm. I... | J Med Chem 49: 2750-7 (2006) Article DOI: 10.1021/jm060077j BindingDB Entry DOI: 10.7270/Q2V98691 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||