

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Wt: 398.4 BDBM28422  Purchase Purchase | Wt: 470.5 BDBM50362000  | Wt: 862.9 BDBM50362001  | Wt: 862.9 BDBM50362002 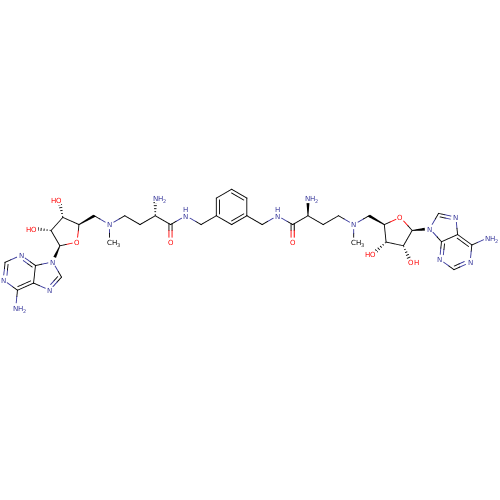 | Wt: 834.8 BDBM50362005 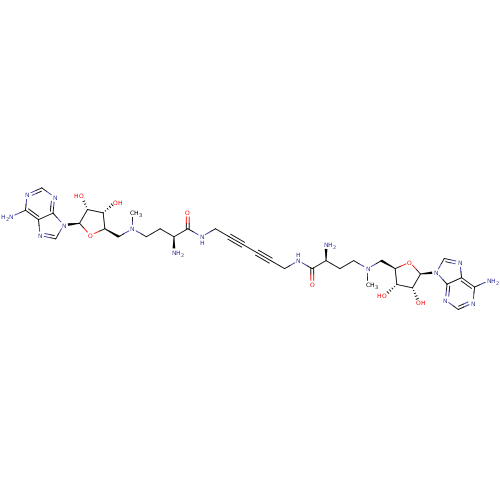 |
| Wt: 485.5 BDBM50362006  | Wt: 893.0 BDBM50362007 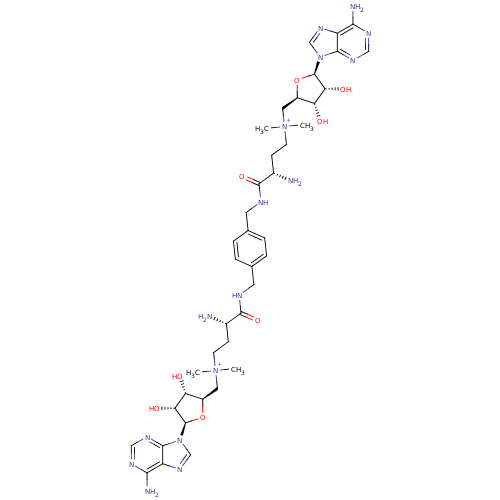 | Wt: 407.4 BDBM50362009  | Wt: 756.8 BDBM50362012 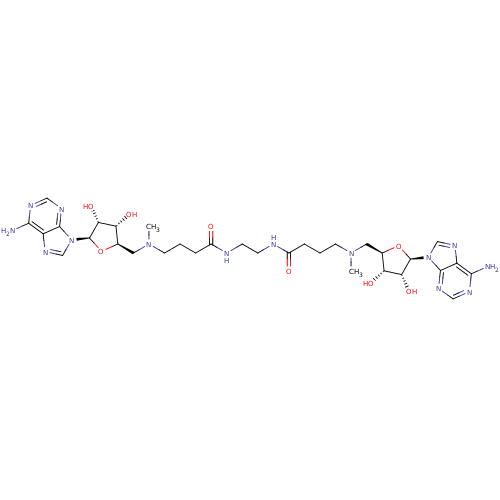 | Wt: 770.8 BDBM50362013  |
| Wt: 784.8 BDBM50362014 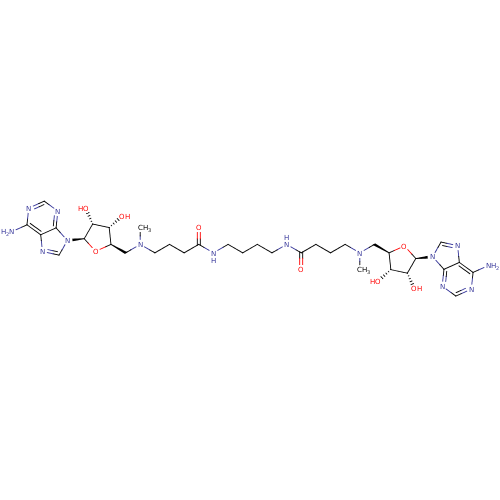 | Wt: 812.9 BDBM50362015  | Wt: 422.5 BDBM50362016 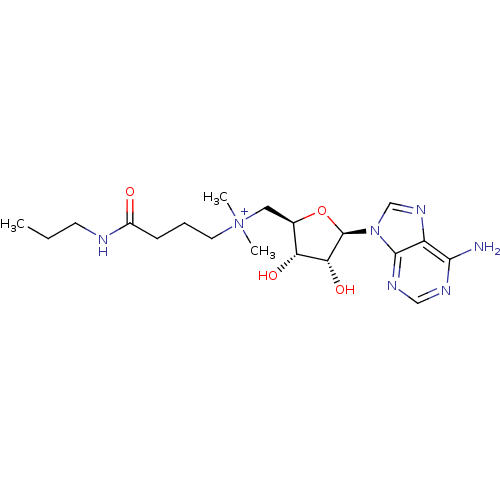 | Wt: 800.9 BDBM50362019  | Wt: 842.9 BDBM50362020 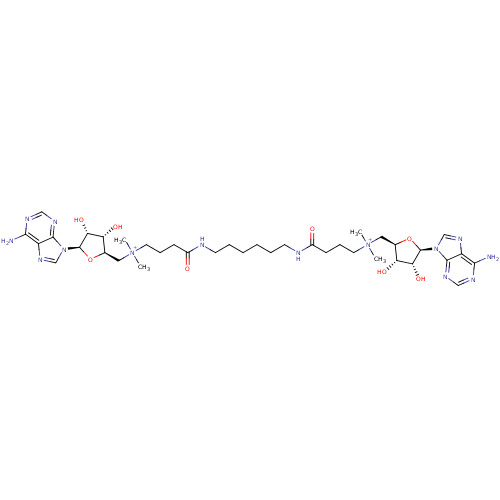 |
| Displayed 1 to 15 (of 22 total ) | Next | Last >> |
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adenosine receptor A1 (Rattus norvegicus (rat)) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 675 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute of Diabetes, Digestive and Kidney Diseases Curated by PDSP Ki Database | Mol Pharmacol 45: 1101-11 (1994) BindingDB Entry DOI: 10.7270/Q26Q1VR9 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Transmembrane and immunoglobulin domain-containing 3 (RAT) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | KEGG UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 2.47E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute of Diabetes, Digestive and Kidney Diseases Curated by PDSP Ki Database | Mol Pharmacol 45: 1101-11 (1994) BindingDB Entry DOI: 10.7270/Q26Q1VR9 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Adenosine receptor A2a (Rattus norvegicus (rat)) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 2.78E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute of Diabetes, Digestive and Kidney Diseases Curated by PDSP Ki Database | Mol Pharmacol 45: 1101-11 (1994) BindingDB Entry DOI: 10.7270/Q26Q1VR9 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-adenosylmethionine decarboxylase proenzyme (Rattus norvegicus) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 6.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Inhibition of H-Ras-mediated farnesylation expressed in mouse NIH3T3 cells | J Med Chem 25: 550-6 (1982) BindingDB Entry DOI: 10.7270/Q2VX0H37 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase (Homo sapiens (Human)) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 1.64E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
C.N.R.S. Curated by ChEMBL | Assay Description Kinetic constant was measured for PMII of Leishmania donovani promastigotes using protein extracted from the 12000 g pellet (P12). | J Med Chem 35: 63-7 (1992) BindingDB Entry DOI: 10.7270/Q2T1548J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase (Homo sapiens (Human)) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 1.64E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
C.N.R.S. Curated by ChEMBL | Assay Description Kinetic constant was measured for PMII of Leishmania donovani promastigotes using protein extracted from the 12000 g pellet (P12). | J Med Chem 35: 63-7 (1992) BindingDB Entry DOI: 10.7270/Q2T1548J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-adenosylmethionine decarboxylase proenzyme (Rattus norvegicus) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PubMed | 3.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description In vitro inhibitory activity against S-adenosyl-L-methionine decarboxylase using liver from rat in absence of putrescine | J Med Chem 25: 550-6 (1982) BindingDB Entry DOI: 10.7270/Q2VX0H37 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protein arginine N-methyltransferase 1 [11-371] (Homo sapiens (Human)) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | 1.20E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
University Park Curated by ChEMBL | Assay Description Inhibition of GST-fused human recombinant PRMT1 after 90 mins by SDS-PAGE based scintillation counting | Bioorg Med Chem Lett 20: 2103-5 (2010) Article DOI: 10.1016/j.bmcl.2010.02.069 BindingDB Entry DOI: 10.7270/Q2WQ04SW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 4.72E+4 | n/a | n/a | n/a | 7.5 | 4 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.91E+4 | n/a | n/a | n/a | 7.5 | 4 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.07E+4 | n/a | n/a | n/a | 7.5 | 4 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 5.80E+3 | n/a | n/a | n/a | 7.5 | 4 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362000 (CHEMBL1939713) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 120 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362001 (CHEMBL1939712) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 61 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362002 (CHEMBL1939711) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 37 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362005 (CHEMBL1939708) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 111 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362006 (CHEMBL1939714) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 36 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362007 (CHEMBL1939715) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 15 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362009 (CHEMBL1939717) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 1.00E+3 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362012 (CHEMBL1939720) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 700 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362013 (CHEMBL1939721) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 320 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362014 (CHEMBL1939722) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 220 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362015 (CHEMBL1939723) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 47 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362016 (CHEMBL1939724) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 150 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362019 (CHEMBL1939727) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 108 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Met repressor (Escherichia coli) | BDBM50362020 (CHEMBL1939728) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | 51 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phenylethanolamine N-methyltransferase (Homo sapiens (Human)) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 4.80E+3 | n/a | n/a | n/a | n/a | n/a |
University of Michigan Curated by ChEMBL | Assay Description Activity of human wild type PNMT assessed as phenylethanolamine methylation | J Med Chem 48: 7243-52 (2005) Article DOI: 10.1021/jm050568o BindingDB Entry DOI: 10.7270/Q2QC049M | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Met repressor (Escherichia coli) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | n/a | 17 | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ assessed as protein dimer-DNA complex formation using F-metC operator DNA by fluorescence anisotropy | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Met repressor (Escherichia coli) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 4 | n/a | n/a | n/a | n/a | n/a |
University of Leeds Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli metJ in presence of operator DNA complex by filter binding study | Bioorg Med Chem Lett 22: 278-84 (2011) Article DOI: 10.1016/j.bmcl.2011.11.017 BindingDB Entry DOI: 10.7270/Q2HQ40BH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 2.67E+4 | n/a | n/a | n/a | 7.5 | 25 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.66E+4 | n/a | n/a | n/a | 7.5 | 20 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 6.10E+3 | n/a | n/a | n/a | 7.5 | 5 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 8.20E+3 | n/a | n/a | n/a | 7.5 | 10 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Phosphoethanolamine N-methyltransferase 1 (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.14E+4 | n/a | n/a | n/a | 7.5 | 15 |
Washington University | Assay Description For standard ITC analysis of ligand binding, proteins were dialyzed overnight in 25 mm Hepes (pH 7.5), 100 mm NaCl, 5 mm ??-mercaptoethanol, and 5% g... | J Biol Chem 286: 38060-8 (2011) Article DOI: 10.1074/jbc.M111.290619 BindingDB Entry DOI: 10.7270/Q2GF0S39 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell (A) | Syringe (B) | Cell Links | Syringe Links | Cell + Syr Links | ΔG° kJ/mole | -TΔS° kJ/mole | ΔH° kJ/mole | log K | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -29 | 23.0 | -53.2 | n/a | 7.5 | 4 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -28 | 0.283 | -28 | n/a | 7.5 | 10 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -28 | -8.34 | -19 | n/a | 7.5 | 5 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -28 | 27.7 | -56.9 | n/a | 7.5 | 4 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -27 | 10.1 | -37 | n/a | 7.5 | 15 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -27 | 19.9 | -46.5 | n/a | 7.5 | 20 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -26 | 26.1 | -52.3 | n/a | 7.5 | 4 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -26 | 32.8 | -59.9 | n/a | 7.5 | 25 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||
| Phosphoehtnaolamine Methyltransferases 1 (PMT1) (Caenorhabditis elegans) | BDBM28422 ((2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-puri...) | GoogleScholar | CHEBI KEGG MMDB PC cid PC sid PDB | -24 | 38.8 | -63.2 | n/a | 7.5 | 4 | |
Washington University | J Biol Chem 286: 38060-8 (2011) | |||||||||