

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26442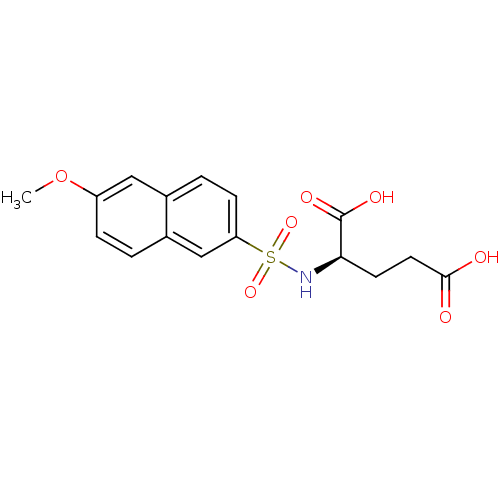 ((2R)-2-[(6-methoxynaphthalene-2-)sulfonamido]penta...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 5.90E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26444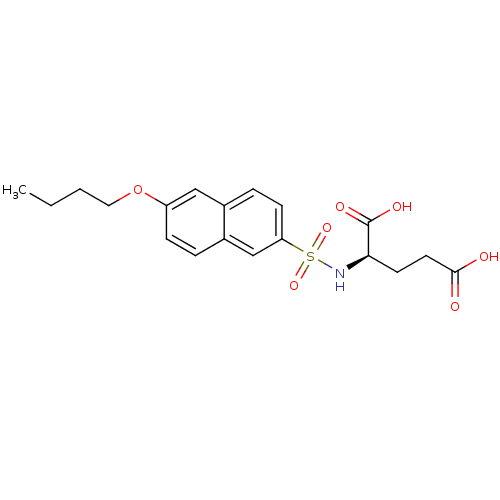 ((2R)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 1.20E+5 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 123 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26444 ((2R)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 2.80E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26445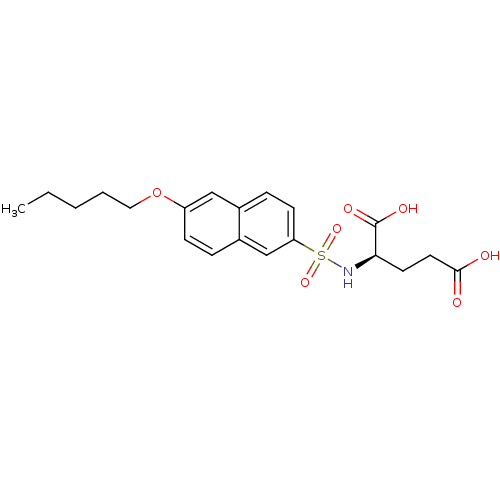 ((2R)-2-{[6-(pentyloxy)naphthalene-2-]sulfonamido}p...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | 1.70E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26446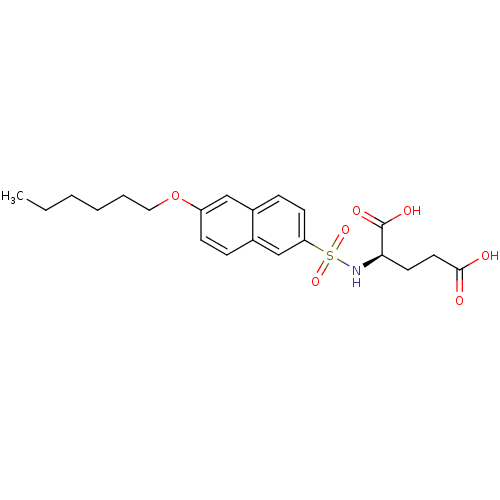 ((2R)-2-{[6-(hexyloxy)naphthalene-2-]sulfonamido}pe...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.76E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26448 ((2R)-2-{[6-(carboxymethoxy)naphthalene-2-]sulfonam...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 6.30E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26449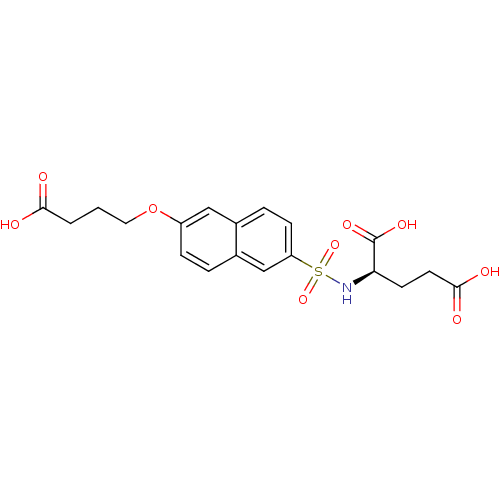 ((2R)-2-{[6-(3-carboxypropoxy)naphthalene-2-]sulfon...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | >1.00E+6 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26450 ((2R)-2-{[6-(cyclopentyloxy)naphthalene-2-]sulfonam...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 4.00E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26459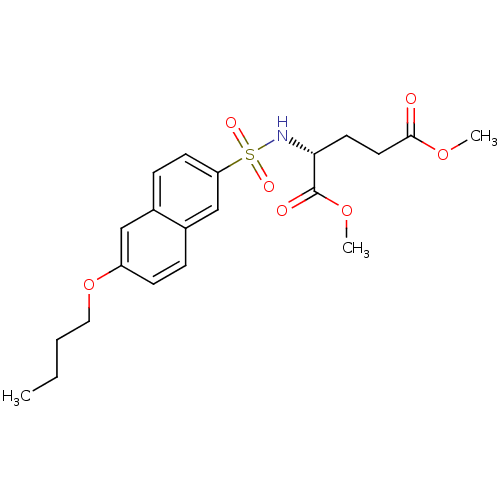 (1,5-dimethyl (2R)-2-[(6-butoxynaphthalene-2-)sulfo...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | >2.00E+6 | n/a | n/a | n/a | n/a | n/a | n/a |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26460 ((2S)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | 7.10E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26463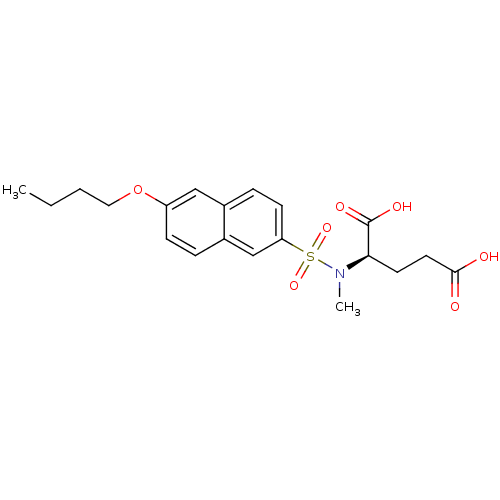 ((2R)-2-[(6-butoxynaphthalene-2-)(methyl)sulfonamid...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | >1.00E+6 | n/a | n/a | n/a | n/a | n/a | n/a |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26465 ((2R)-2-[(5-butoxynaphthalene-1-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | >1.00E+6 | n/a | n/a | n/a | n/a | n/a | n/a |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26466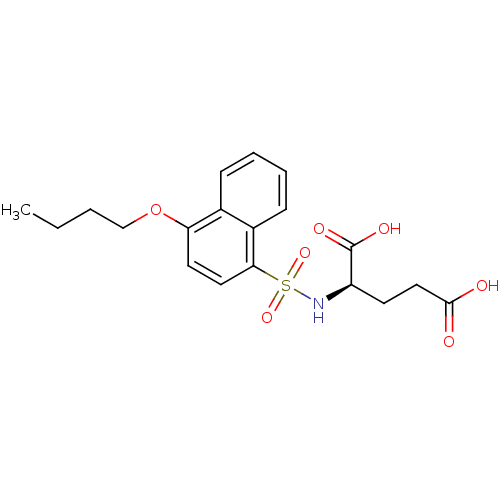 ((2R)-2-[(4-butoxynaphthalene-1-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | >1.00E+6 | n/a | n/a | n/a | n/a | n/a | n/a |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26444 ((2R)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 2.80E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia | Bioorg Chem 55: 2-15 (2014) Article DOI: 10.1016/j.bioorg.2014.03.008 BindingDB Entry DOI: 10.7270/Q2ZG6QWR | ||||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26460 ((2S)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | 7.10E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana, A?kerceva 7, 1000 Ljubljana, Slovenia | Bioorg Chem 55: 2-15 (2014) Article DOI: 10.1016/j.bioorg.2014.03.008 BindingDB Entry DOI: 10.7270/Q2ZG6QWR | ||||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26444 ((2R)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 2.10E+5 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 4 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26443 ((2R)-2-[(6-propoxynaphthalene-2-)sulfonamido]penta...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.05E+5 | n/a | n/a | n/a | n/a | 8.6 | 37 |
Lek Pharmaceuticals d.d. | Assay Description The compounds were tested for their ability to inhibit the addition of D-[14C]Glu to UMA. The radioactive substrate and product were separated by re... | J Med Chem 51: 7486-94 (2008) Article DOI: 10.1021/jm800762u BindingDB Entry DOI: 10.7270/Q2B27SMN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||