

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein mono-ADP-ribosyltransferase PARP15 [460-656,Y598L] (Homo sapiens (Human)) | BDBM199181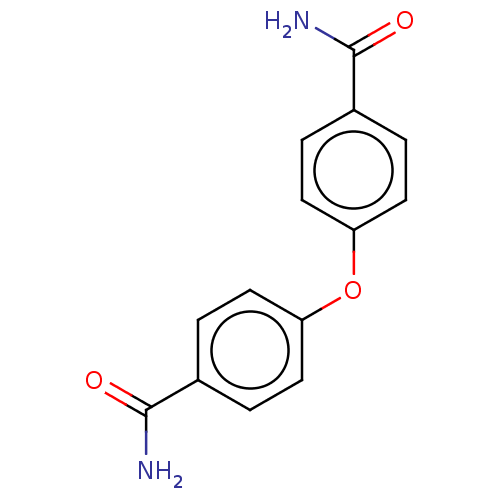 (4-[(4-Carbamoylcyclohexyl)oxy]cyclohexane-1-carbox...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 295 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Oulu | Assay Description This assay measures the substrate consumption by quantifying the remaining NAD+ through its chemical conversion to a stable fluorescent condensation ... | Cell Chem Biol 23: 1251-1260 (2016) Article DOI: 10.1016/j.chembiol.2016.08.012 BindingDB Entry DOI: 10.7270/Q2B56HJ3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||