 Found 5 hits of kd for monomerid = 20732
Found 5 hits of kd for monomerid = 20732 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Heat shock protein HSP 90-beta
(Homo sapiens (Human)) | BDBM20732
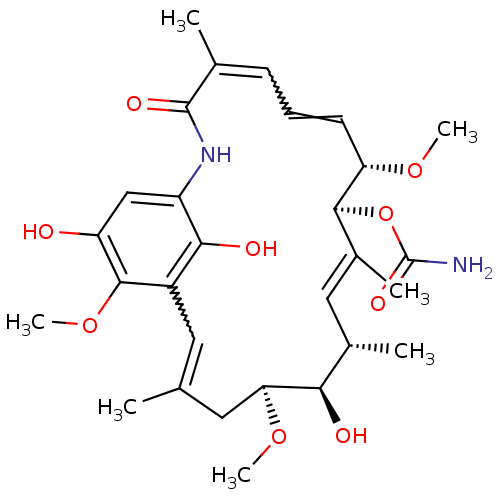
((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14,...)Show SMILES CO[C@H]1CC(C)=Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2OC |r,w:16.16,6.6,t:28| Show InChI InChI=1S/C29H40N2O9/c1-15-11-19-25(34)20(14-21(32)27(19)39-7)31-28(35)16(2)9-8-10-22(37-5)26(40-29(30)36)18(4)13-17(3)24(33)23(12-15)38-6/h8-11,13-14,17,22-24,26,32-34H,12H2,1-7H3,(H2,30,36)(H,31,35)/b10-8?,15-11?,16-9?,18-13+/t17-,22-,23-,24+,26-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 670 | n/a | n/a | n/a | n/a | n/a |
Kosan Biosciences, Inc.
Curated by ChEMBL
| Assay Description
Binding affinity to human recombinant HSP90 |
J Med Chem 52: 3265-73 (2009)
Article DOI: 10.1021/jm900098v
BindingDB Entry DOI: 10.7270/Q2V40W5R |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Heat shock protein HSP 90-alpha
(Homo sapiens (Human)) | BDBM20732

((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14,...)Show SMILES CO[C@H]1CC(C)=Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2OC |r,w:16.16,6.6,t:28| Show InChI InChI=1S/C29H40N2O9/c1-15-11-19-25(34)20(14-21(32)27(19)39-7)31-28(35)16(2)9-8-10-22(37-5)26(40-29(30)36)18(4)13-17(3)24(33)23(12-15)38-6/h8-11,13-14,17,22-24,26,32-34H,12H2,1-7H3,(H2,30,36)(H,31,35)/b10-8?,15-11?,16-9?,18-13+/t17-,22-,23-,24+,26-/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 775 | n/a | n/a | n/a | n/a | n/a |
University College London
Curated by ChEMBL
| Assay Description
The compound was tested for binding affinity against isolated N-domain of Heat shock protein HSP 90 |
J Med Chem 42: 260-6 (1999)
Article DOI: 10.1021/jm980403y
BindingDB Entry DOI: 10.7270/Q29G5NHQ |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Heat shock protein HSP 90-beta
(Homo sapiens (Human)) | BDBM20732

((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14,...)Show SMILES CO[C@H]1CC(C)=Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2OC |r,w:16.16,6.6,t:28| Show InChI InChI=1S/C29H40N2O9/c1-15-11-19-25(34)20(14-21(32)27(19)39-7)31-28(35)16(2)9-8-10-22(37-5)26(40-29(30)36)18(4)13-17(3)24(33)23(12-15)38-6/h8-11,13-14,17,22-24,26,32-34H,12H2,1-7H3,(H2,30,36)(H,31,35)/b10-8?,15-11?,16-9?,18-13+/t17-,22-,23-,24+,26-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 1.00E+3 | n/a | n/a | n/a | n/a | n/a |
San Diego State University
Curated by ChEMBL
| Assay Description
Binding affinity to Hsp90 N-domain |
Bioorg Med Chem Lett 21: 7068-71 (2011)
Article DOI: 10.1016/j.bmcl.2011.09.096
BindingDB Entry DOI: 10.7270/Q22R3S3T |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
ATP-dependent molecular chaperone HSP82
(Saccharomyces cerevisiae) | BDBM20732

((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14,...)Show SMILES CO[C@H]1CC(C)=Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2OC |r,w:16.16,6.6,t:28| Show InChI InChI=1S/C29H40N2O9/c1-15-11-19-25(34)20(14-21(32)27(19)39-7)31-28(35)16(2)9-8-10-22(37-5)26(40-29(30)36)18(4)13-17(3)24(33)23(12-15)38-6/h8-11,13-14,17,22-24,26,32-34H,12H2,1-7H3,(H2,30,36)(H,31,35)/b10-8?,15-11?,16-9?,18-13+/t17-,22-,23-,24+,26-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | n/a | 1.20E+3 | n/a | n/a | n/a | n/a | n/a |
Memorial Sloan-Kettering Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to N-terminal ATP/ADP-binding domain of yeast Hsp90 |
Bioorg Med Chem 17: 2225-35 (2009)
Article DOI: 10.1016/j.bmc.2008.10.087
BindingDB Entry DOI: 10.7270/Q2T72JCS |
More data for this
Ligand-Target Pair | 
3D Structure (docked) |
Heat shock protein HSP 90-alpha
(Homo sapiens (Human)) | BDBM20732

((4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14,...)Show SMILES CO[C@H]1CC(C)=Cc2c(O)c(NC(=O)C(C)=CC=C[C@H](OC)[C@@H](OC(N)=O)\C(C)=C\[C@H](C)[C@H]1O)cc(O)c2OC |r,w:16.16,6.6,t:28| Show InChI InChI=1S/C29H40N2O9/c1-15-11-19-25(34)20(14-21(32)27(19)39-7)31-28(35)16(2)9-8-10-22(37-5)26(40-29(30)36)18(4)13-17(3)24(33)23(12-15)38-6/h8-11,13-14,17,22-24,26,32-34H,12H2,1-7H3,(H2,30,36)(H,31,35)/b10-8?,15-11?,16-9?,18-13+/t17-,22-,23-,24+,26-/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 1.22E+3 | n/a | n/a | n/a | n/a | n/a |
University College London
Curated by ChEMBL
| Assay Description
The compound was tested for binding affinity against yeast Heat shock protein HSP 90 |
J Med Chem 42: 260-6 (1999)
Article DOI: 10.1021/jm980403y
BindingDB Entry DOI: 10.7270/Q29G5NHQ |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data