

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 protease (Human immunodeficiency virus) | BDBM104103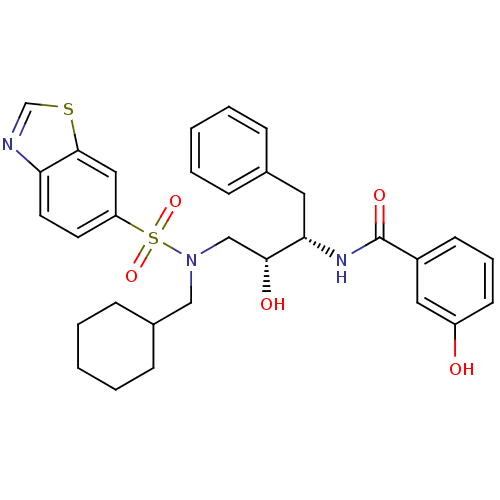 (N-[(1S,2R)-3-[(6-Benzothiazolylsulfonyl)(cyclohexy...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.129 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Massachusetts Institute of Technology | Assay Description HIV protease inhibitor activities were determined by fluorescence resonance energy transfer (FRET) method. Protease substrate, Arg-Glu-(EDANS)-Ser-G... | ACS Chem Biol 8: 2433-41 (2013) Article DOI: 10.1021/cb400468c BindingDB Entry DOI: 10.7270/Q2R2101Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,I494L,I497V,V499I,D514N,E519D,R541K,D544E,Q553K,N572D,L573M] (Human immunodeficiency virus) | BDBM104103 (N-[(1S,2R)-3-[(6-Benzothiazolylsulfonyl)(cyclohexy...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 0.383 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Massachusetts Institute of Technology | Assay Description HIV protease inhibitor activities were determined by fluorescence resonance energy transfer (FRET) method. Protease substrate, Arg-Glu-(EDANS)-Ser-G... | ACS Chem Biol 8: 2433-41 (2013) Article DOI: 10.1021/cb400468c BindingDB Entry DOI: 10.7270/Q2R2101Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,I497V,V499I,E519D,G532V,I538V,R541K,D544E,Q553K,V566A,L573M] (Human immunodeficiency virus) | BDBM104103 (N-[(1S,2R)-3-[(6-Benzothiazolylsulfonyl)(cyclohexy...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.33 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Massachusetts Institute of Technology | Assay Description HIV protease inhibitor activities were determined by fluorescence resonance energy transfer (FRET) method. Protease substrate, Arg-Glu-(EDANS)-Ser-G... | ACS Chem Biol 8: 2433-41 (2013) Article DOI: 10.1021/cb400468c BindingDB Entry DOI: 10.7270/Q2R2101Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,I497V,V499I,E519D,R541K,D544E,Q553K,A555V,G557S,I568V,L574M,L573M] (Human immunodeficiency virus) | BDBM104103 (N-[(1S,2R)-3-[(6-Benzothiazolylsulfonyl)(cyclohexy...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 2.75 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Massachusetts Institute of Technology | Assay Description HIV protease inhibitor activities were determined by fluorescence resonance energy transfer (FRET) method. Protease substrate, Arg-Glu-(EDANS)-Ser-G... | ACS Chem Biol 8: 2433-41 (2013) Article DOI: 10.1021/cb400468c BindingDB Entry DOI: 10.7270/Q2R2101Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,I494L,I497V,V499I,E519D,I534V,R541K,D544E,Q553K,A555V,L573M] (Human immunodeficiency virus) | BDBM104103 (N-[(1S,2R)-3-[(6-Benzothiazolylsulfonyl)(cyclohexy...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 4.82 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Massachusetts Institute of Technology | Assay Description HIV protease inhibitor activities were determined by fluorescence resonance energy transfer (FRET) method. Protease substrate, Arg-Glu-(EDANS)-Ser-G... | ACS Chem Biol 8: 2433-41 (2013) Article DOI: 10.1021/cb400468c BindingDB Entry DOI: 10.7270/Q2R2101Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||