

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heat shock 70 kDa protein 1A (Homo sapiens (Human)) | BDBM32378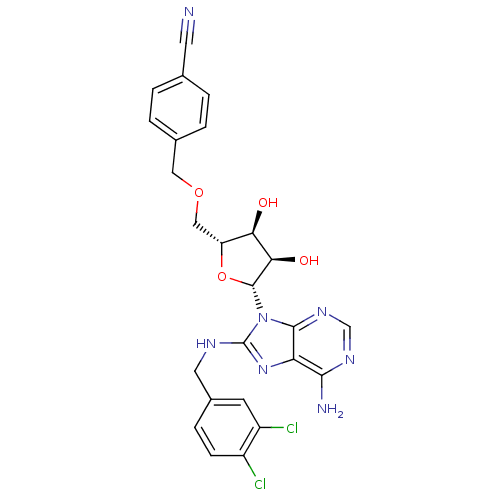 (adenosine-derived inhibitor (Grp78), 13 | adenosin...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase MCE MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 120 | -39.5 | n/a | n/a | n/a | n/a | n/a | 7.4 | 25 |
Vernalis (R&D) Ltd. | Assay Description The assay was based on the fluorescence polarization of the unbound small binding partner will be low, and its binding to a larger binding partner wi... | J Med Chem 54: 4034-41 (2011) Article DOI: 10.1021/jm101625x BindingDB Entry DOI: 10.7270/Q2R49P83 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Heat shock-related 70 kDa protein 2 (Homo sapiens) | BDBM32378 (adenosine-derived inhibitor (Grp78), 13 | adenosin...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | >200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research Curated by ChEMBL | Assay Description Covalent inhibition of HSP72-NBD (unknown origin) nucleotide-derived FP probe displacement based by fluorescence polarization assay | J Med Chem 62: 11383-11398 (2019) Article DOI: 10.1021/acs.jmedchem.9b01709 BindingDB Entry DOI: 10.7270/Q22R3W5Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Heat shock-related 70 kDa protein 2 (Homo sapiens) | BDBM32378 (adenosine-derived inhibitor (Grp78), 13 | adenosin...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | <200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research Curated by ChEMBL | Assay Description Covalent inhibition of HSP72-NBD (unknown origin) nucleotide-derived FP probe displacement based by fluorescence polarization assay | J Med Chem 62: 11383-11398 (2019) Article DOI: 10.1021/acs.jmedchem.9b01709 BindingDB Entry DOI: 10.7270/Q22R3W5Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Heat shock-related 70 kDa protein 2 (Homo sapiens) | BDBM32378 (adenosine-derived inhibitor (Grp78), 13 | adenosin...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | <200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research Curated by ChEMBL | Assay Description Covalent inhibition of HSP72-NBD (unknown origin) nucleotide-derived FP probe displacement based by fluorescence polarization assay | J Med Chem 62: 11383-11398 (2019) Article DOI: 10.1021/acs.jmedchem.9b01709 BindingDB Entry DOI: 10.7270/Q22R3W5Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Heat shock-related 70 kDa protein 2 (Homo sapiens) | BDBM32378 (adenosine-derived inhibitor (Grp78), 13 | adenosin...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | >200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research Curated by ChEMBL | Assay Description Covalent inhibition of HSP72-NBD (unknown origin) nucleotide-derived FP probe displacement based by fluorescence polarization assay | J Med Chem 62: 11383-11398 (2019) Article DOI: 10.1021/acs.jmedchem.9b01709 BindingDB Entry DOI: 10.7270/Q22R3W5Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||