 Found 2 hits of ki for monomerid = 50078165
Found 2 hits of ki for monomerid = 50078165 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Induced myeloid leukemia cell differentiation protein Mcl-1
(Homo sapiens (Human)) | BDBM50078165
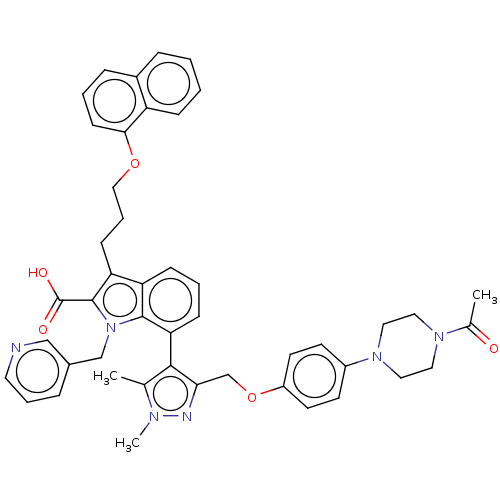
(CHEMBL3417706)Show SMILES CC(=O)N1CCN(CC1)c1ccc(OCc2nn(C)c(C)c2-c2cccc3c(CCCOc4cccc5ccccc45)c(C(O)=O)n(Cc4cccnc4)c23)cc1 |(19.93,14.21,;19.31,15.27,;19.92,16.34,;17.77,15.26,;16.99,16.59,;15.45,16.59,;14.69,15.25,;15.46,13.92,;17,13.93,;13.15,15.25,;12.38,13.91,;10.84,13.91,;10.07,15.24,;8.52,15.23,;7.76,13.9,;6.22,13.89,;5.3,15.13,;3.84,14.63,;2.84,15.35,;3.86,13.09,;2.87,12.35,;5.32,12.66,;5.8,11.19,;7.33,10.86,;7.8,9.4,;6.76,8.24,;5.25,8.57,;4.01,7.7,;4.01,6.16,;2.67,5.39,;2.67,3.85,;1.33,3.08,;1.33,1.54,;2.66,.77,;2.66,-.77,;1.33,-1.54,;,-.77,;-1.33,-1.54,;-2.68,-.77,;-2.68,.77,;-1.33,1.54,;,.77,;2.78,8.56,;1.33,8.05,;.4,8.86,;1.09,6.84,;3.25,10.03,;2.34,11.27,;.8,11.1,;.19,9.69,;-1.34,9.51,;-2.26,10.75,;-1.64,12.16,;-.11,12.34,;4.78,10.03,;10.83,16.58,;12.37,16.58,)| Show InChI InChI=1S/C46H46N6O5/c1-31-43(41(48-49(31)3)30-57-36-20-18-35(19-21-36)51-25-23-50(24-26-51)32(2)53)40-15-7-14-38-39(16-9-27-56-42-17-6-12-34-11-4-5-13-37(34)42)45(46(54)55)52(44(38)40)29-33-10-8-22-47-28-33/h4-8,10-15,17-22,28H,9,16,23-27,29-30H2,1-3H3,(H,54,55) | PDB
MMDB
NCI pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 1.80 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc.
Curated by ChEMBL
| Assay Description
Displacement of F-Bak (GQVGRQLAIIGDK(6-FAM)INR-amide) from MCL1 (unknown origin) after 1 hr by TR-FRET assay |
J Med Chem 58: 2180-94 (2015)
Article DOI: 10.1021/jm501258m
BindingDB Entry DOI: 10.7270/Q2HX1FC8 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Induced myeloid leukemia cell differentiation protein Mcl-1
(Homo sapiens (Human)) | BDBM50078165

(CHEMBL3417706)Show SMILES CC(=O)N1CCN(CC1)c1ccc(OCc2nn(C)c(C)c2-c2cccc3c(CCCOc4cccc5ccccc45)c(C(O)=O)n(Cc4cccnc4)c23)cc1 |(19.93,14.21,;19.31,15.27,;19.92,16.34,;17.77,15.26,;16.99,16.59,;15.45,16.59,;14.69,15.25,;15.46,13.92,;17,13.93,;13.15,15.25,;12.38,13.91,;10.84,13.91,;10.07,15.24,;8.52,15.23,;7.76,13.9,;6.22,13.89,;5.3,15.13,;3.84,14.63,;2.84,15.35,;3.86,13.09,;2.87,12.35,;5.32,12.66,;5.8,11.19,;7.33,10.86,;7.8,9.4,;6.76,8.24,;5.25,8.57,;4.01,7.7,;4.01,6.16,;2.67,5.39,;2.67,3.85,;1.33,3.08,;1.33,1.54,;2.66,.77,;2.66,-.77,;1.33,-1.54,;,-.77,;-1.33,-1.54,;-2.68,-.77,;-2.68,.77,;-1.33,1.54,;,.77,;2.78,8.56,;1.33,8.05,;.4,8.86,;1.09,6.84,;3.25,10.03,;2.34,11.27,;.8,11.1,;.19,9.69,;-1.34,9.51,;-2.26,10.75,;-1.64,12.16,;-.11,12.34,;4.78,10.03,;10.83,16.58,;12.37,16.58,)| Show InChI InChI=1S/C46H46N6O5/c1-31-43(41(48-49(31)3)30-57-36-20-18-35(19-21-36)51-25-23-50(24-26-51)32(2)53)40-15-7-14-38-39(16-9-27-56-42-17-6-12-34-11-4-5-13-37(34)42)45(46(54)55)52(44(38)40)29-33-10-8-22-47-28-33/h4-8,10-15,17-22,28H,9,16,23-27,29-30H2,1-3H3,(H,54,55) | PDB
MMDB
NCI pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 98 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc.
Curated by ChEMBL
| Assay Description
Displacement of F-Bak (GQVGRQLAIIGDK(6-FAM)INR-amide) from MCL1 (unknown origin) after 1 hr by TR-FRET assay in presence of 10% human serum |
J Med Chem 58: 2180-94 (2015)
Article DOI: 10.1021/jm501258m
BindingDB Entry DOI: 10.7270/Q2HX1FC8 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data



