 Found 10 hits Enz. Inhib. hit(s) with Target = 'E3 ubiquitin-protein ligase Mdm2' and Ligand = 'BDBM50433561'
Found 10 hits Enz. Inhib. hit(s) with Target = 'E3 ubiquitin-protein ligase Mdm2' and Ligand = 'BDBM50433561' Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561
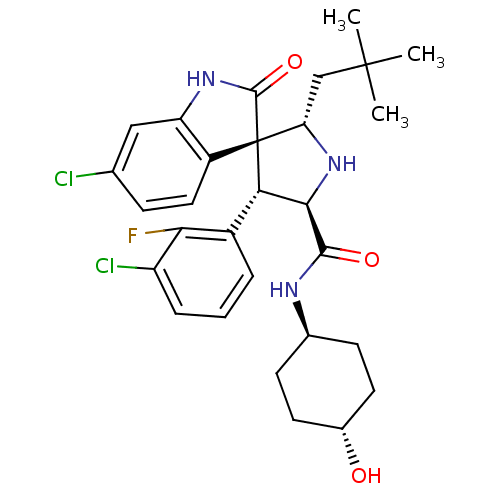
(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Michigan Comprehensive Cancer Center and Departments of Internal Medicine
Curated by ChEMBL
| Assay Description
Binding affinity to MDM2 (unknown origin) |
J Med Chem 58: 1038-52 (2015)
Article DOI: 10.1021/jm501092z
BindingDB Entry DOI: 10.7270/Q21G0NZ5 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
JSS Academy of Higher Education and Research
Curated by ChEMBL
| Assay Description
Inhibition of FAM-tagged p53-based fluorescent probe binding to human His-tagged MDM2 (1 to 118 residues) after 15 mins by fluorescence polarization ... |
Eur J Med Chem 158: 7-24 (2018)
Article DOI: 10.1016/j.ejmech.2018.08.099
BindingDB Entry DOI: 10.7270/Q28G8PC5 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Inhibition of FAM tagged p53-based peptide binding to recombinant human His-tagged MDM2 protein (residues 1 to 118 residues) incubated for 15 mins by... |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.0c00829
BindingDB Entry DOI: 10.7270/Q22J6GHJ |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Wisconsin-Madison
Curated by ChEMBL
| Assay Description
Binding affinity to MDM2 (unknown origin) |
Eur J Med Chem 176: 476-491 (2019)
Article DOI: 10.1016/j.ejmech.2019.05.046
BindingDB Entry DOI: 10.7270/Q22J6G6H |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sichuan University and Collaborative Innovation Center for Biotherapy
Curated by ChEMBL
| Assay Description
Inhibition of PMDM6-F binding to recombinant human His-tagged MDM2 (1 to 118 residues) after 15 mins by fluorescence polarization assay |
Eur J Med Chem 126: 1071-1082 (2017)
Article DOI: 10.1016/j.ejmech.2016.12.021
BindingDB Entry DOI: 10.7270/Q2JW8H51 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Binding affinity to recombinant human His6-tagged HDM2 (1 to 118 residues) assessed as reduction in PMDM6-F binding incubated for 15 to 30 mins by fl... |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.1c01524
BindingDB Entry DOI: 10.7270/Q2Z03D1V |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 0.880 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
International Institute for Translational Chinese Medicine
Curated by ChEMBL
| Assay Description
Inhibition of MDM2 (unknown origin) |
Eur J Med Chem 159: 1-9 (2018)
Article DOI: 10.1016/j.ejmech.2018.09.044
BindingDB Entry DOI: 10.7270/Q2SB48FV |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| 10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanofi
Curated by ChEMBL
| Assay Description
Binding affinity to human MDM2 |
Bioorg Med Chem Lett 23: 2480-5 (2013)
Article DOI: 10.1016/j.bmcl.2013.03.034
BindingDB Entry DOI: 10.7270/Q2RN3970 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | 100 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Inhibition of MDM2 (unknown origin) by cell-based assay |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.0c00829
BindingDB Entry DOI: 10.7270/Q22J6GHJ |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(Homo sapiens (Human)) | BDBM50433561

(CHEMBL2381408)Show SMILES CC(C)(C)C[C@@H]1N[C@H]([C@H](c2cccc(Cl)c2F)[C@]11C(=O)Nc2cc(Cl)ccc12)C(=O)N[C@H]1CC[C@H](O)CC1 |r,wU:8.8,5.4,31.34,wD:17.29,7.31,34.38,(40.88,-57.18,;39.34,-57.18,;38.57,-58.51,;40.11,-58.5,;38.57,-55.84,;37.04,-55.84,;37.52,-54.37,;36.27,-53.47,;35.03,-54.37,;33.51,-54.09,;32.52,-55.27,;31.01,-54.99,;30.49,-53.53,;31.49,-52.36,;30.98,-50.9,;33,-52.64,;34.01,-51.48,;35.49,-55.84,;36.38,-57.1,;37.93,-57.11,;35.48,-58.34,;34.01,-57.86,;32.69,-58.61,;31.36,-57.84,;30.03,-58.6,;31.37,-56.3,;32.71,-55.54,;34.03,-56.32,;36.26,-51.93,;34.93,-51.16,;37.6,-51.15,;37.6,-49.61,;36.26,-48.86,;36.26,-47.31,;37.6,-46.54,;37.6,-45,;38.93,-47.32,;38.93,-48.85,)| Show InChI InChI=1S/C29H34Cl2FN3O3/c1-28(2,3)14-22-29(19-12-7-15(30)13-21(19)34-27(29)38)23(18-5-4-6-20(31)24(18)32)25(35-22)26(37)33-16-8-10-17(36)11-9-16/h4-7,12-13,16-17,22-23,25,35-36H,8-11,14H2,1-3H3,(H,33,37)(H,34,38)/t16-,17-,22-,23-,25+,29+/m0/s1 | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 2.70 | n/a | n/a | n/a | n/a | n/a |
Amgen Inc.
Curated by ChEMBL
| Assay Description
Binding affinity to human MDM2 by by Surface Plasmon Resonace (SPR) spectroscopy binding assay |
J Med Chem 57: 1454-72 (2014)
Article DOI: 10.1021/jm401753e
BindingDB Entry DOI: 10.7270/Q24M960Z |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data