

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mu-type opioid receptor (Homo sapiens (Human)) | BDBM50087016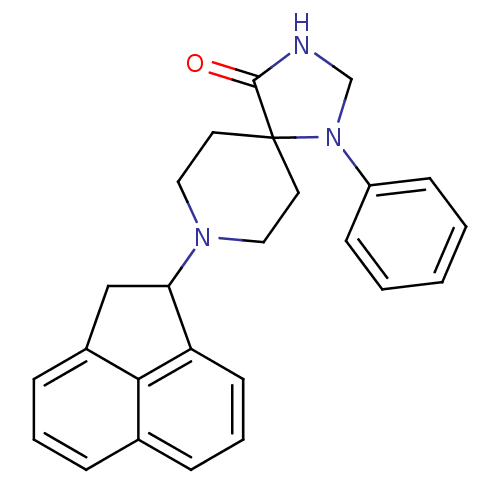 (8-Acenaphthen-1-yl-1-phenyl-1,3,8-triaza-spiro[4.5...) | PDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE PC cid PC sid UniChem Patents Similars | PubMed | 5.90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
F. Hoffmann-La Roche Ltd. Curated by ChEMBL | Assay Description Competitive binding displacement analyses was performed from permanently transfected HEK293 cells expressing human Opioid receptor mu 1 | J Med Chem 43: 1329-38 (2001) BindingDB Entry DOI: 10.7270/Q2ZG6SZG | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Homo sapiens (Human)) | BDBM50087016 (8-Acenaphthen-1-yl-1-phenyl-1,3,8-triaza-spiro[4.5...) | PDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE PC cid PC sid UniChem Patents Similars | Article PubMed | 12.9 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Leicester Royal Infirmary Curated by PDSP Ki Database | Naunyn Schmiedebergs Arch Pharmacol 363: 28-33 (2001) Article DOI: 10.1007/s002100000327 BindingDB Entry DOI: 10.7270/Q2TH8K8W | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Homo sapiens (Human)) | BDBM50087016 (8-Acenaphthen-1-yl-1-phenyl-1,3,8-triaza-spiro[4.5...) | PDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE PC cid PC sid UniChem Patents Similars | PubMed | n/a | n/a | n/a | n/a | 2.50E+3 | n/a | n/a | n/a | n/a |
F. Hoffmann-La Roche Ltd. Curated by ChEMBL | Assay Description Effective concentration of the required to stimulate binding of GTPgammaS to mu1 receptor was determined using scintillation proximity assay | J Med Chem 43: 1329-38 (2001) BindingDB Entry DOI: 10.7270/Q2ZG6SZG | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||