

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate--tRNA ligase (Escherichia coli) | BDBM18122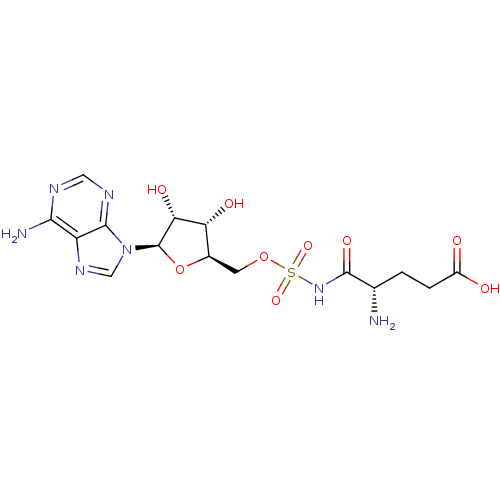 ((4S)-4-amino-5-[({[(2R,3S,4R,5R)-5-(6-amino-9H-pur...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 2.80 | -12.1 | n/a | n/a | n/a | n/a | n/a | 7.2 | 37 |
CREFSIP | Assay Description The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p... | J Enzyme Inhib Med Chem 20: 61-7 (2005) Article DOI: 10.1080/14756360400002007 BindingDB Entry DOI: 10.7270/Q2X0659C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate--tRNA ligase (Escherichia coli) | BDBM18123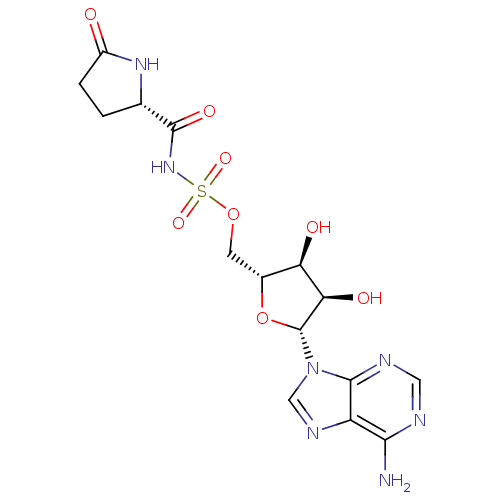 ((5S)-5-{[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 1.50E+4 | -6.84 | n/a | n/a | n/a | n/a | n/a | 7.2 | 37 |
CREFSIP | Assay Description The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p... | J Enzyme Inhib Med Chem 20: 61-7 (2005) Article DOI: 10.1080/14756360400002007 BindingDB Entry DOI: 10.7270/Q2X0659C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate--tRNA ligase (Escherichia coli) | BDBM18118 ((4S)-4-amino-6-({[(2R,3S,4R,5R)-5-(6-amino-9H-puri...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 1.80E+4 | -6.73 | n/a | n/a | n/a | n/a | n/a | 7.2 | 37 |
CREFSIP | Assay Description The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p... | Bioorg Med Chem 15: 295-304 (2007) Article DOI: 10.1016/j.bmc.2006.09.056 BindingDB Entry DOI: 10.7270/Q21N7ZD3 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate--tRNA ligase (Escherichia coli) | BDBM18120 (Gln-KPA | [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 2.90E+6 | -3.60 | n/a | n/a | n/a | n/a | n/a | 7.2 | 37 |
CREFSIP | Assay Description The Km and Kappm for the amino acid substrate were first calculated from Lineweaver-Burk plots. The Ki values were calculated from the Kappm vs [I] p... | Bioorg Med Chem 15: 295-304 (2007) Article DOI: 10.1016/j.bmc.2006.09.056 BindingDB Entry DOI: 10.7270/Q21N7ZD3 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||