

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31657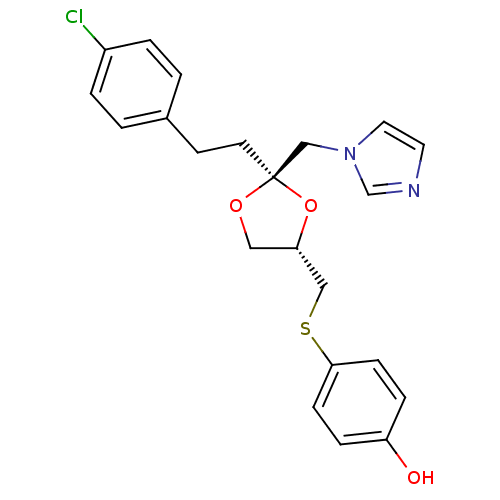 (imidazole-dioxolane, 9) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31665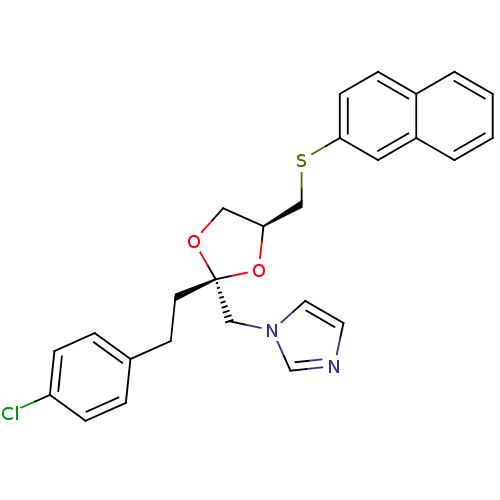 (imidazole-dioxolane, 17) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31652 (imidazole-dioxolane, 3) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31669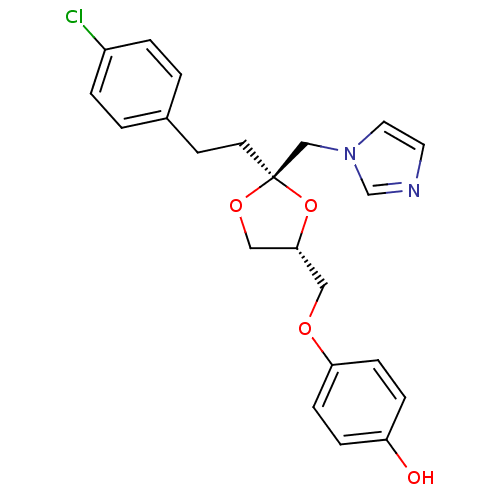 (imidazole-dioxolane, 21) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31658 (imidazole-dioxolane, 10) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31681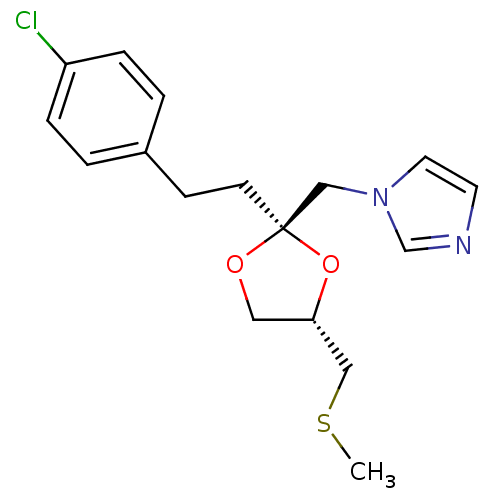 (imidazole-dioxolane, 31) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31664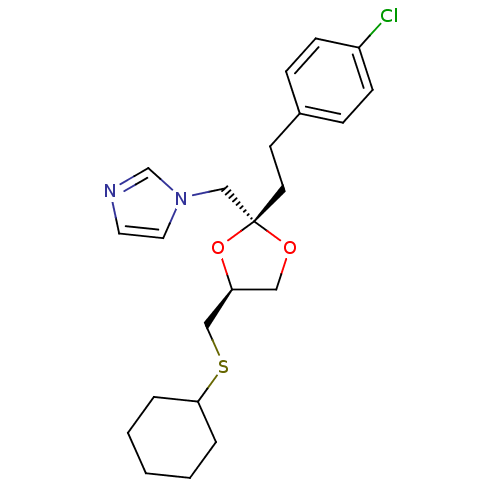 (imidazole-dioxolane, 16) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31672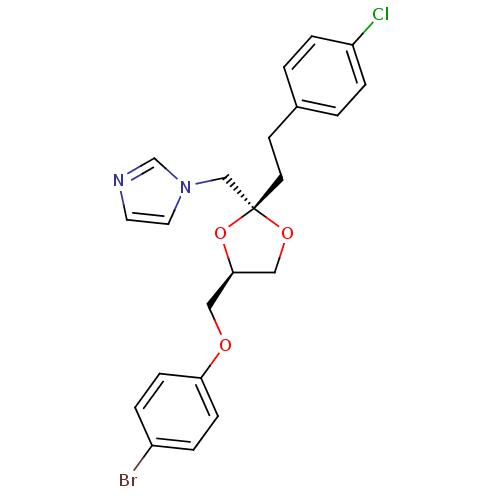 (imidazole-dioxolane, 24) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 3.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31682 (imidazole-dioxolane, 32) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 4.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31677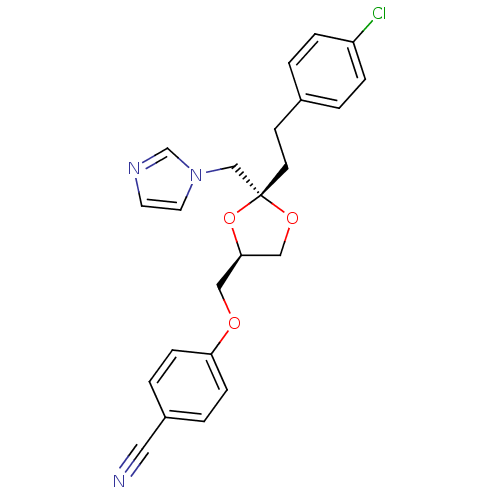 (imidazole-dioxolane, 29) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 6.90E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM31656 (imidazole-dioxolane, 8) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 7.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Queen's University | Assay Description CYP3A1/3A2-catalyzed erythromycin N-demethylase activity was determined by the spectrophotometric measurement of formaldehyde. Briefly, reaction mixt... | Bioorg Med Chem 17: 2461-75 (2009) Article DOI: 10.1016/j.bmc.2009.01.078 BindingDB Entry DOI: 10.7270/Q2WQ0250 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cytochrome P450 3A1 (Rattus norvegicus (rat)) | BDBM261607 (US10513499, Compound 18 | US11254644, Compound 18 ...) | UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | Purchase MCE PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | >1.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
TES Pharma S.r.l. Curated by ChEMBL | Assay Description Inhibition of recombinant rat CYP3A1 expressed in baculosomes expression system using fluorogenic-BOMCC as substrate by fluorescent homogeneous assay | J Med Chem 61: 745-759 (2018) Article DOI: 10.1021/acs.jmedchem.7b01254 BindingDB Entry DOI: 10.7270/Q22J6F80 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||