

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lysosomal alpha-glucosidase (Mus musculus (Mouse)) | BDBM243075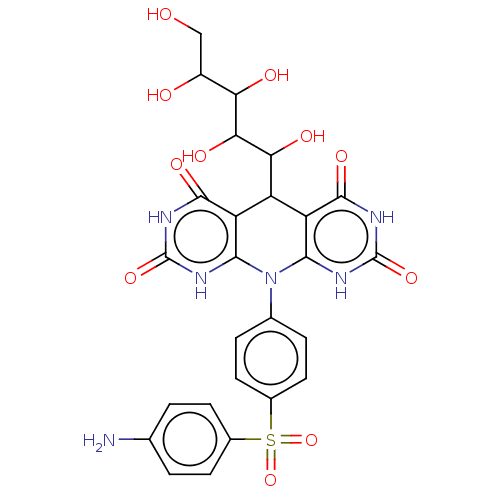 (α-Gl inhibitor, C3) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 2.50E+3 | n/a | n/a | n/a | n/a | n/a | n/a | 7.0 | n/a |
Shiraz University | Assay Description In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth... | J Enzyme Inhib Med Chem 28: 1228-35 (2013) Article DOI: 10.3109/14756366.2012.727812 BindingDB Entry DOI: 10.7270/Q2XP73V0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84968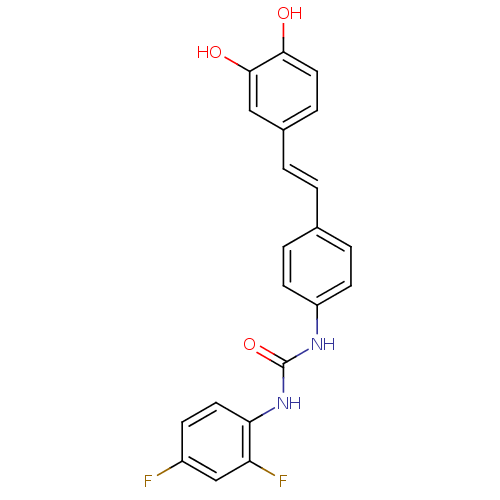 (Urea derivative, 12) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 3.20E+3 | n/a | 8.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84969 (Urea derivative, 13) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 4.60E+3 | n/a | 1.43E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84967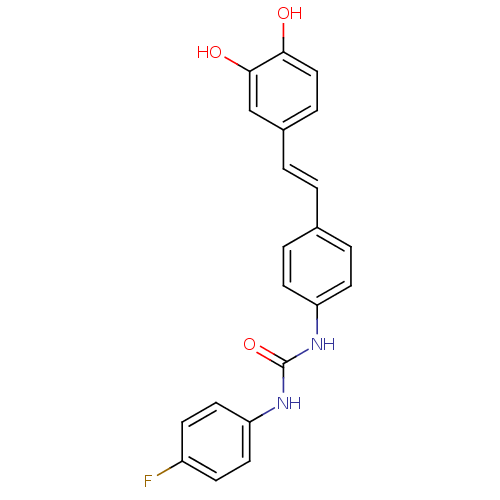 (Urea derivative, 11) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 7.20E+3 | n/a | 1.98E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84972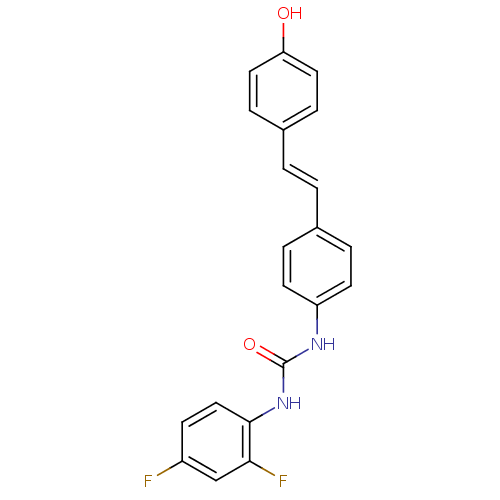 (Urea derivative, 16) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 9.40E+3 | n/a | 1.85E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84963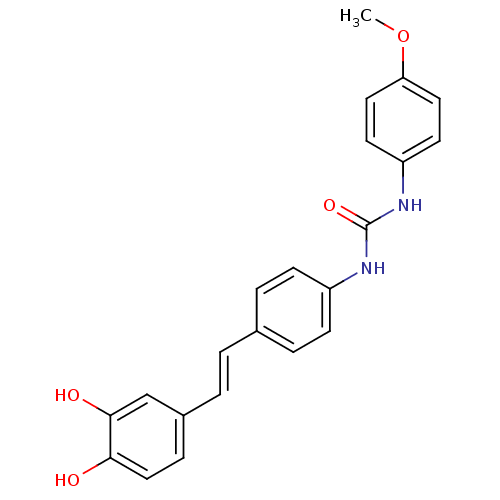 (Urea derivative, 7) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 1.05E+4 | n/a | 1.95E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84962 (Urea derivative, 6) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 1.06E+4 | n/a | 4.21E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84966 (Urea derivative, 10) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 1.21E+4 | n/a | 2.88E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM243075 (α-Gl inhibitor, C3) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 1.40E+4 | n/a | n/a | n/a | n/a | n/a | n/a | 7.0 | n/a |
Shiraz University | Assay Description In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth... | J Enzyme Inhib Med Chem 28: 1228-35 (2013) Article DOI: 10.3109/14756366.2012.727812 BindingDB Entry DOI: 10.7270/Q2XP73V0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84970 (Urea derivative, 14) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 1.70E+4 | n/a | 2.91E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM18351 ((2R,3R,4R,5S)-2-(Hydroxymethyl)piperidine-3,4,5-tr...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 1.80E+4 | n/a | 3.95E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM23926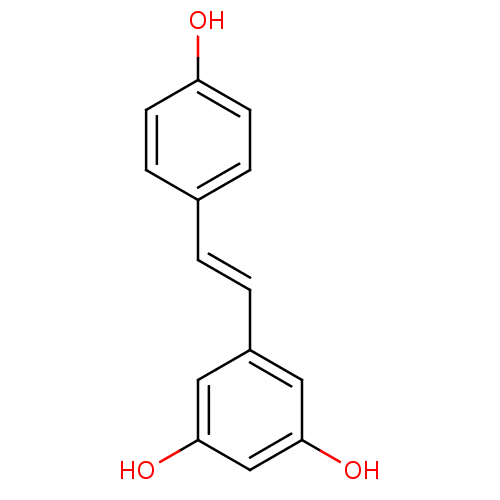 ((E)-resveratrol | 5-[(E)-2-(4-hydroxyphenyl)etheny...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 2.15E+4 | n/a | 2.64E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM243074 (α-Gl inhibitor, C2) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 3.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | 7.0 | n/a |
Shiraz University | Assay Description In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth... | J Enzyme Inhib Med Chem 28: 1228-35 (2013) Article DOI: 10.3109/14756366.2012.727812 BindingDB Entry DOI: 10.7270/Q2XP73V0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM84959 (Stilbene derivative, 2) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 3.51E+4 | n/a | 4.64E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Gyeongsang National University | Assay Description All enzymatic activities were determined by using the appropriate substrate (p-nitrophenyl-alpha-D-glucopyranoside, p-nitrophenyl-beta-D-gulcopyranos... | Chembiochem 11: 2125-31 (2010) Article DOI: 10.1002/cbic.201000376 BindingDB Entry DOI: 10.7270/Q2542M33 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM243073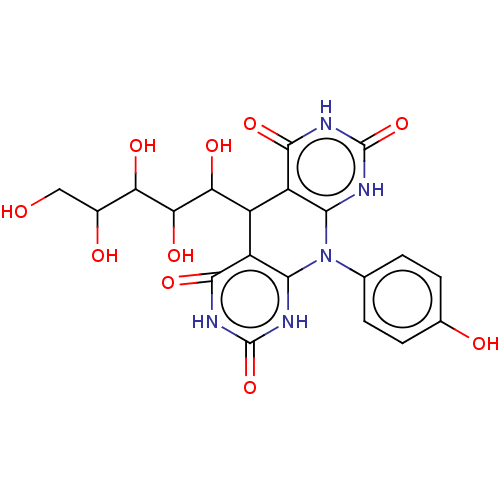 (α-Gl inhibitor, C1) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 7.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | 7.0 | n/a |
Shiraz University | Assay Description In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth... | J Enzyme Inhib Med Chem 28: 1228-35 (2013) Article DOI: 10.3109/14756366.2012.727812 BindingDB Entry DOI: 10.7270/Q2XP73V0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM243074 (α-Gl inhibitor, C2) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 1.30E+5 | n/a | n/a | n/a | n/a | n/a | n/a | 7.0 | n/a |
Shiraz University | Assay Description In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth... | J Enzyme Inhib Med Chem 28: 1228-35 (2013) Article DOI: 10.3109/14756366.2012.727812 BindingDB Entry DOI: 10.7270/Q2XP73V0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-glucosidase MAL12 (Saccharomyces cerevisiae) | BDBM243073 (α-Gl inhibitor, C1) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 1.70E+5 | n/a | n/a | n/a | n/a | n/a | n/a | 7.0 | n/a |
Shiraz University | Assay Description In this study, the inhibition assay of yeast enzyme was performed in 100 mM phosphate buffer pH 7.0 at 25°C with minor changes, according to the meth... | J Enzyme Inhib Med Chem 28: 1228-35 (2013) Article DOI: 10.3109/14756366.2012.727812 BindingDB Entry DOI: 10.7270/Q2XP73V0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||