

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rho-associated protein kinase 1 (Homo sapiens (Human)) | BDBM14028 ((S)-2-METHYL-1-[(4-METHYL-5-ISOQUINOLINE)SULFONYL]...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents | DrugBank PDB Article PubMed | 6 | -47.7 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Rho-associated protein kinase 1 (Homo sapiens (Human)) | BDBM14031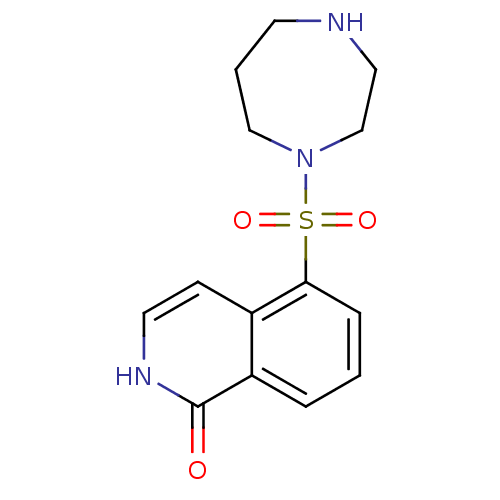 (5-(1,4-diazepane-1-sulfonyl)-1,2-dihydroisoquinoli...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 150 | -39.6 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Rho-associated protein kinase 1 (Homo sapiens (Human)) | BDBM14029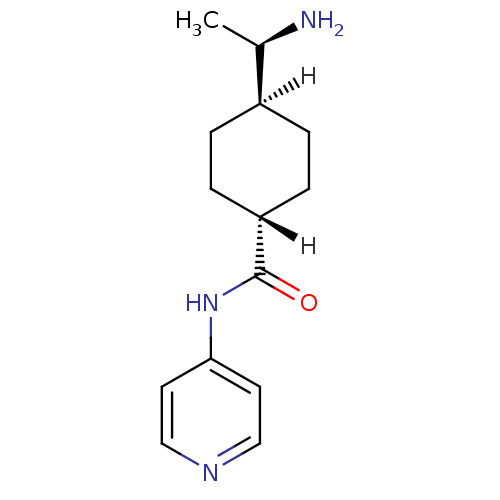 ((R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXA...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank MMDB PDB Article PubMed | 150 | -39.6 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| cAMP-dependent protein kinase catalytic subunit alpha (Mus musculus (mouse)) | BDBM14028 ((S)-2-METHYL-1-[(4-METHYL-5-ISOQUINOLINE)SULFONYL]...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents | PDB Article PubMed | 340 | -37.5 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| cAMP-dependent protein kinase catalytic subunit alpha (Mus musculus (mouse)) | BDBM14027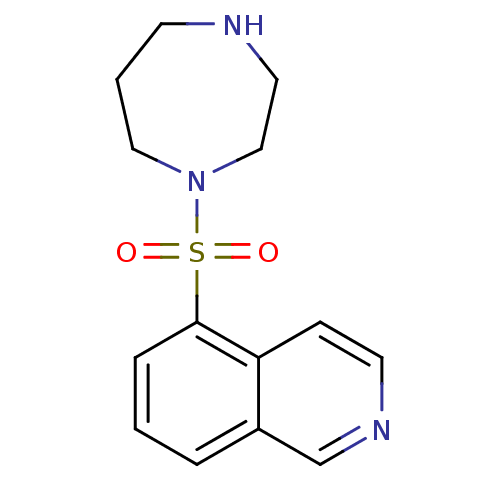 (5-(1,4-diazepan-1-ylsulfonyl)isoquinoline | 5-(1,4...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 460 | -36.8 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Rho-associated protein kinase 1 (Homo sapiens (Human)) | BDBM14027 (5-(1,4-diazepan-1-ylsulfonyl)isoquinoline | 5-(1,4...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | 530 | -36.4 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| cAMP-dependent protein kinase catalytic subunit alpha (Mus musculus (mouse)) | BDBM14031 (5-(1,4-diazepane-1-sulfonyl)-1,2-dihydroisoquinoli...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 2.20E+3 | -32.8 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| cAMP-dependent protein kinase catalytic subunit alpha (Mus musculus (mouse)) | BDBM14029 ((R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXA...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | >5.00E+3 | >-30.8 | n/a | n/a | n/a | n/a | n/a | 7.6 | 30 |
Vertex Pharmaceuticals | Assay Description The enzyme activity was assayed by using an ATP regenerative NADH consuming system. The reaction was started with adding ATP to the mixture containin... | J Biol Chem 281: 260-8 (2006) Article DOI: 10.1074/jbc.M508847200 BindingDB Entry DOI: 10.7270/Q2JQ0Z80 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||