

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26455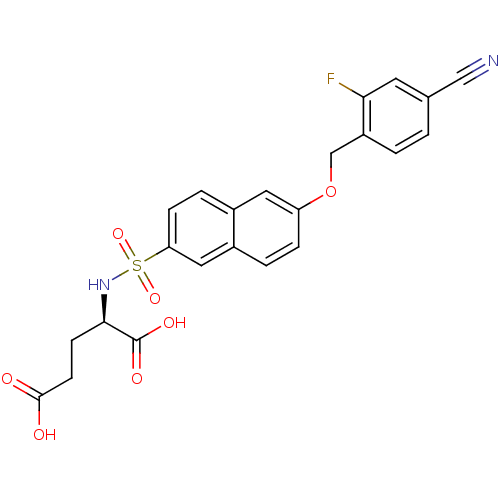 ((2R)-2-({6-[(4-cyano-2-fluorophenyl)methoxy]naphth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | n/a | 2.20E+3 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 9 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26455 ((2R)-2-({6-[(4-cyano-2-fluorophenyl)methoxy]naphth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | n/a | 5.50E+3 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 56 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26455 ((2R)-2-({6-[(4-cyano-2-fluorophenyl)methoxy]naphth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | n/a | 7.00E+3 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 13 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM50021359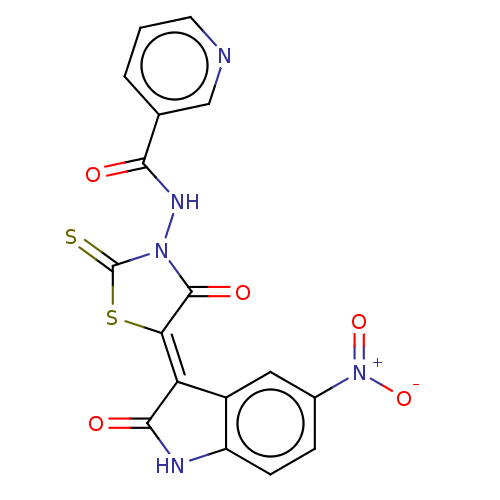 (CHEMBL3288597) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 7.90E+4 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 43 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM50021359 (CHEMBL3288597) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 9.80E+4 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM50021359 (CHEMBL3288597) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.06E+5 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 71 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM50021359 (CHEMBL3288597) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.11E+5 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 4 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26444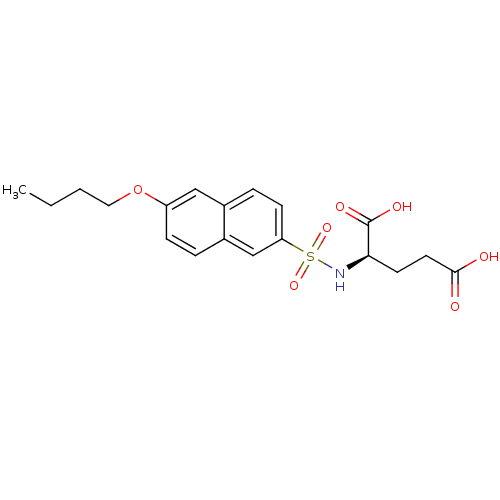 ((2R)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 1.20E+5 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 123 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| UDP-N-acetylmuramoylalanine--D-glutamate ligase (Escherichia coli (strain K12)) | BDBM26444 ((2R)-2-[(6-butoxynaphthalene-2-)sulfonamido]pentan...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 2.10E+5 | n/a | n/a | n/a | n/a | n/a |
Slovenia National Institute of Chemistry Curated by ChEMBL | Assay Description Binding affinity to 6x His-tagged and 15N/13C-labeled Escherichia coli MurD ligase measured at NMR signal 4 by 1H/13C-HSQC 2D NMR spectroscopy | Eur J Med Chem 83: 92-101 (2014) Article DOI: 10.1016/j.ejmech.2014.06.021 BindingDB Entry DOI: 10.7270/Q25D8TDN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||