 Found 12 hits of kd data for polymerid = 50004421
Found 12 hits of kd data for polymerid = 50004421 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50595370
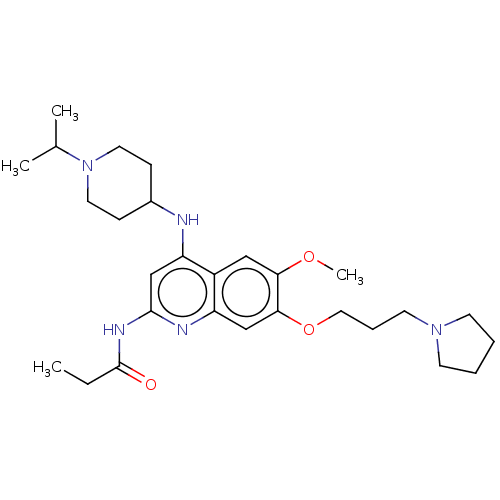
(CHEMBL5189737)Show SMILES CCC(=O)Nc1cc(NC2CCN(CC2)C(C)C)c2cc(OC)c(OCCCN3CCCC3)cc2n1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 16 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00652
BindingDB Entry DOI: 10.7270/Q26T0RPT |
More data for this
Ligand-Target Pair | |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50300032
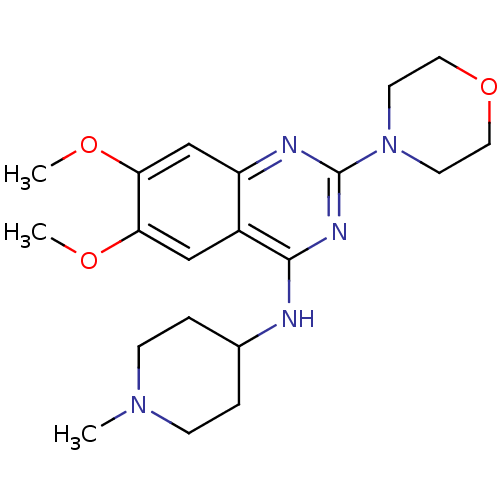
(6,7-Dimethoxy-N-(1-methylpiperidin-4-yl)-2-morphol...)Show InChI InChI=1S/C20H29N5O3/c1-24-6-4-14(5-7-24)21-19-15-12-17(26-2)18(27-3)13-16(15)22-20(23-19)25-8-10-28-11-9-25/h12-14H,4-11H2,1-3H3,(H,21,22,23) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 40 | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Binding affinity to recombinant GLP catalytic SET domain (982 to 1266 residues) (unknown origin) expressed in Escherichia coli BL21 (DE3) codon plus ... |
J Med Chem 60: 1876-1891 (2017)
Article DOI: 10.1021/acs.jmedchem.6b01645
BindingDB Entry DOI: 10.7270/Q2ZP4947 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50501525
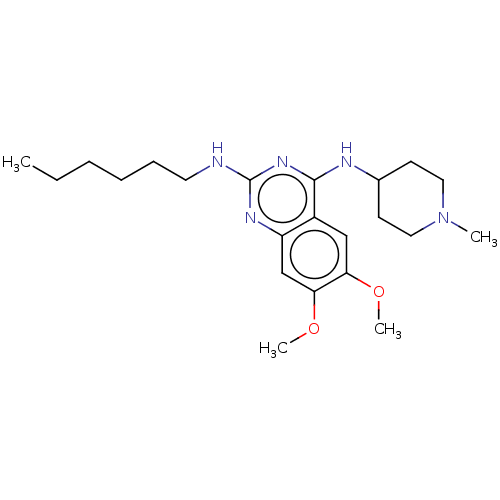
(CHEMBL4086403)Show SMILES CCCCCCNc1nc(NC2CCN(C)CC2)c2cc(OC)c(OC)cc2n1 Show InChI InChI=1S/C22H35N5O2/c1-5-6-7-8-11-23-22-25-18-15-20(29-4)19(28-3)14-17(18)21(26-22)24-16-9-12-27(2)13-10-16/h14-16H,5-13H2,1-4H3,(H2,23,24,25,26) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 46 | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Binding affinity to recombinant GLP catalytic SET domain (982 to 1266 residues) (unknown origin) expressed in Escherichia coli BL21 (DE3) codon plus ... |
J Med Chem 60: 1876-1891 (2017)
Article DOI: 10.1021/acs.jmedchem.6b01645
BindingDB Entry DOI: 10.7270/Q2ZP4947 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50595369
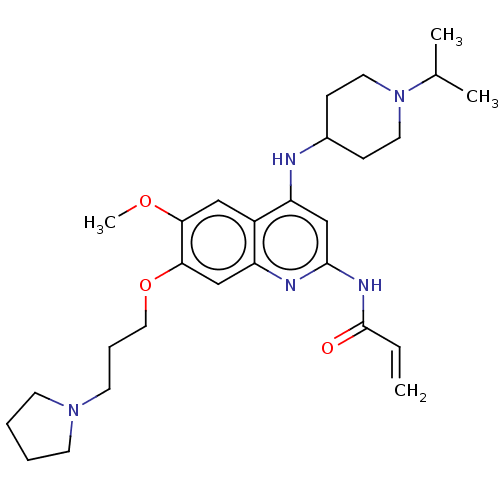
(CHEMBL5170450)Show SMILES COc1cc2c(NC3CCN(CC3)C(C)C)cc(NC(=O)C=C)nc2cc1OCCCN1CCCC1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 46 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00652
BindingDB Entry DOI: 10.7270/Q26T0RPT |
More data for this
Ligand-Target Pair | |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50268886
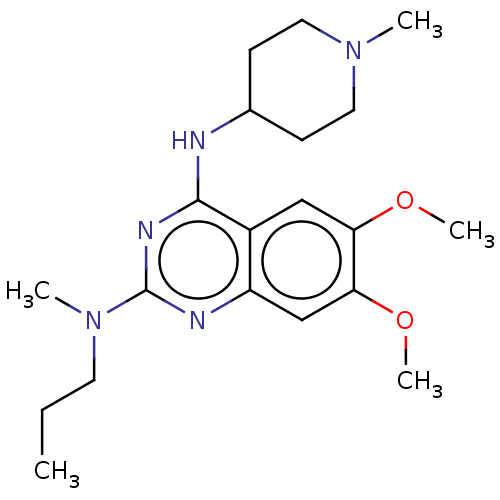
(CHEMBL4072211)Show SMILES CCCN(C)c1nc(NC2CCN(C)CC2)c2cc(OC)c(OC)cc2n1 Show InChI InChI=1S/C20H31N5O2/c1-6-9-25(3)20-22-16-13-18(27-5)17(26-4)12-15(16)19(23-20)21-14-7-10-24(2)11-8-14/h12-14H,6-11H2,1-5H3,(H,21,22,23) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 57 | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Binding affinity to human GLP in presence of SAM by isothermal titration calorimetry |
Bioorg Med Chem 25: 4414-4423 (2017)
Article DOI: 10.1016/j.bmc.2017.06.021
BindingDB Entry DOI: 10.7270/Q2X350ZG |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50442103
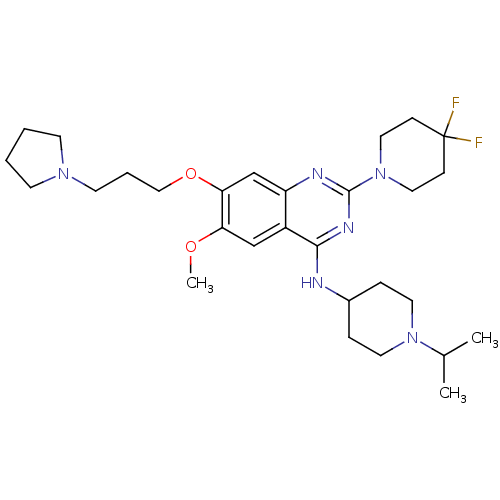
(CHEMBL2441082)Show SMILES COc1cc2c(NC3CCN(CC3)C(C)C)nc(nc2cc1OCCCN1CCCC1)N1CCC(F)(F)CC1 Show InChI InChI=1S/C29H44F2N6O2/c1-21(2)36-14-7-22(8-15-36)32-27-23-19-25(38-3)26(39-18-6-13-35-11-4-5-12-35)20-24(23)33-28(34-27)37-16-9-29(30,31)10-17-37/h19-22H,4-18H2,1-3H3,(H,32,33,34) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | n/a | 62 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00652
BindingDB Entry DOI: 10.7270/Q26T0RPT |
More data for this
Ligand-Target Pair | |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50268879
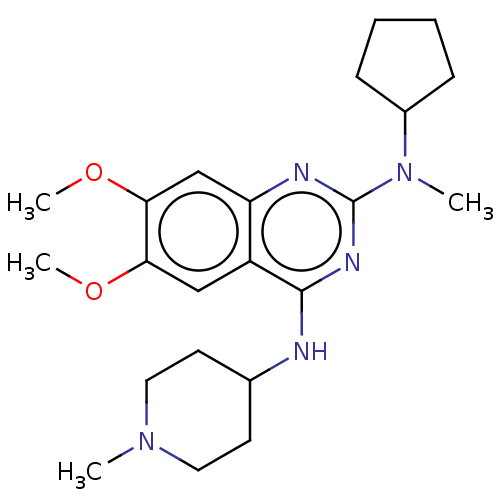
(CHEMBL4094717)Show SMILES COc1cc2nc(nc(NC3CCN(C)CC3)c2cc1OC)N(C)C1CCCC1 Show InChI InChI=1S/C22H33N5O2/c1-26-11-9-15(10-12-26)23-21-17-13-19(28-3)20(29-4)14-18(17)24-22(25-21)27(2)16-7-5-6-8-16/h13-16H,5-12H2,1-4H3,(H,23,24,25) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 94 | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Binding affinity to human GLP in presence of SAM by isothermal titration calorimetry |
Bioorg Med Chem 25: 4414-4423 (2017)
Article DOI: 10.1016/j.bmc.2017.06.021
BindingDB Entry DOI: 10.7270/Q2X350ZG |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50396024
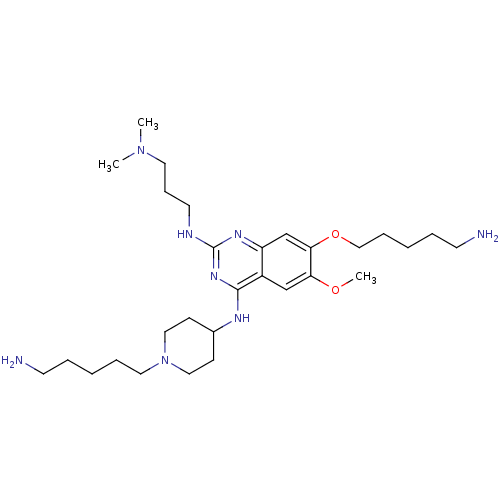
(CHEMBL1232453)Show SMILES COc1cc2c(NC3CCN(CCCCCN)CC3)nc(NCCCN(C)C)nc2cc1OCCCCCN Show InChI InChI=1S/C29H52N8O2/c1-36(2)16-10-15-32-29-34-25-22-27(39-20-9-5-7-14-31)26(38-3)21-24(25)28(35-29)33-23-11-18-37(19-12-23)17-8-4-6-13-30/h21-23H,4-20,30-31H2,1-3H3,(H2,32,33,34,35) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 136 | n/a | n/a | n/a | n/a | n/a |
TBA
Curated by ChEMBL
| Assay Description
Binding affinity to human recombinant GLP catalytic domain amino acid 951 to 1235 expressed in Escherichia coli BL21 (DE3) by isothermal titration ca... |
Eur J Med Chem 56: 179-194 (2012)
Article DOI: 10.1016/j.ejmech.2012.08.010
BindingDB Entry DOI: 10.7270/Q2TQ62NX |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50396024

(CHEMBL1232453)Show SMILES COc1cc2c(NC3CCN(CCCCCN)CC3)nc(NCCCN(C)C)nc2cc1OCCCCCN Show InChI InChI=1S/C29H52N8O2/c1-36(2)16-10-15-32-29-34-25-22-27(39-20-9-5-7-14-31)26(38-3)21-24(25)28(35-29)33-23-11-18-37(19-12-23)17-8-4-6-13-30/h21-23H,4-20,30-31H2,1-3H3,(H2,32,33,34,35) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | n/a | 136 | n/a | n/a | n/a | n/a | n/a |
Icahn School of Medicine at Mount Sinai
Curated by ChEMBL
| Assay Description
Binding affinity to recombinant human GLP (951 to 1235) by isothermal titration calorimetric analysis |
J Med Chem 58: 1596-629 (2015)
Article DOI: 10.1021/jm501234a
BindingDB Entry DOI: 10.7270/Q28K7BS2 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50595368

(CHEMBL5191615)Show SMILES COc1cc2c(NC3CCN(CC3)C(C)C)nc(NC(=O)C=C)nc2cc1OCCCN1CCCC1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 790 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.2c00652
BindingDB Entry DOI: 10.7270/Q26T0RPT |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50282113
(CHEMBL4168535)Show SMILES CNCCCC[C@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=O)[C@@H](NC(=O)[C@H](CCC(N)=O)NC(=O)[C@H](CCCCN)NC(=O)[C@@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=S)Nc1ccc(C2=C3C=CC(O)=CC3Oc3cc(O)ccc23)c(c1)C(O)=O)[C@@H](C)O)[C@@H](C)O)C(=O)N[C@@H](CO)C(=O)N[C@@H]([C@@H](C)O)C(=O)NCC(=O)NCC(=O)N[C@@H](CCCCN)C(=O)N[C@@H](C)C(O)=O |r,c:73,75,78| Show InChI InChI=1S/C85H132N26O26S/c1-40(69(121)103-56(19-14-32-94-83(89)90)73(125)105-54(18-10-13-31-93-7)74(126)108-59(39-112)77(129)109-66(43(4)113)78(130)97-37-63(119)96-38-64(120)102-53(16-8-11-29-86)71(123)99-42(3)81(133)134)98-79(131)67(44(5)114)110-76(128)58(27-28-62(88)118)106-72(124)55(17-9-12-30-87)107-80(132)68(45(6)115)111-75(127)57(20-15-33-95-84(91)92)104-70(122)41(2)100-85(138)101-46-21-24-49(52(34-46)82(135)136)65-50-25-22-47(116)35-60(50)137-61-36-48(117)23-26-51(61)65/h21-26,34-36,40-45,53-60,66-68,93,112-117H,8-20,27-33,37-39,86-87H2,1-7H3,(H2,88,118)(H,96,119)(H,97,130)(H,98,131)(H,99,123)(H,102,120)(H,103,121)(H,104,122)(H,105,125)(H,106,124)(H,107,132)(H,108,126)(H,109,129)(H,110,128)(H,111,127)(H,133,134)(H,135,136)(H4,89,90,94)(H4,91,92,95)(H2,100,101,138)/t40-,41-,42-,43+,44+,45+,53-,54-,55-,56-,57-,58-,59-,60?,66-,67-,68-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| KEGG
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | n/a | 5.00E+3 | n/a | n/a | n/a | n/a | n/a |
University of Connecticut
Curated by ChEMBL
| Assay Description
Binding affinity to GLP (unknown origin) after 30 mins by fluorescence polarization assay |
Eur J Med Chem 136: 14-35 (2017)
Article DOI: 10.1016/j.ejmech.2017.04.047
BindingDB Entry DOI: 10.7270/Q2F76G3G |
More data for this
Ligand-Target Pair | |
Histone-lysine N-methyltransferase EHMT1
(Homo sapiens (Human)) | BDBM50282160
(CHEMBL4176399)Show SMILES C[C@@H](O)[C@H](NC(=O)[C@H](CO)NC(=O)[C@H](CCCCN(C)C)NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=O)[C@@H](NC(=O)[C@H](CCC(N)=O)NC(=O)[C@H](CCCCN)NC(=O)[C@@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=S)Nc1ccc(C2=C3C=CC(O)=CC3Oc3cc(O)ccc23)c(c1)C(O)=O)[C@@H](C)O)[C@@H](C)O)C(=O)NCC(=O)NCC(=O)N[C@@H](CCCCN)C(=O)N[C@@H](C)C(O)=O |r,c:87,89,92| Show InChI InChI=1S/C86H134N26O26S/c1-41(70(122)103-57(20-15-32-94-84(90)91)74(126)105-55(19-11-14-34-112(7)8)75(127)108-60(40-113)78(130)109-67(44(4)114)79(131)97-38-64(120)96-39-65(121)102-54(17-9-12-30-87)72(124)99-43(3)82(134)135)98-80(132)68(45(5)115)110-77(129)59(28-29-63(89)119)106-73(125)56(18-10-13-31-88)107-81(133)69(46(6)116)111-76(128)58(21-16-33-95-85(92)93)104-71(123)42(2)100-86(139)101-47-22-25-50(53(35-47)83(136)137)66-51-26-23-48(117)36-61(51)138-62-37-49(118)24-27-52(62)66/h22-27,35-37,41-46,54-61,67-69,113-118H,9-21,28-34,38-40,87-88H2,1-8H3,(H2,89,119)(H,96,120)(H,97,131)(H,98,132)(H,99,124)(H,102,121)(H,103,122)(H,104,123)(H,105,126)(H,106,125)(H,107,133)(H,108,127)(H,109,130)(H,110,129)(H,111,128)(H,134,135)(H,136,137)(H4,90,91,94)(H4,92,93,95)(H2,100,101,139)/t41-,42-,43-,44+,45+,46+,54-,55-,56-,57-,58-,59-,60-,61?,67-,68-,69-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| KEGG
PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | n/a | 7.00E+3 | n/a | n/a | n/a | n/a | n/a |
University of Connecticut
Curated by ChEMBL
| Assay Description
Binding affinity to GLP (unknown origin) after 30 mins by fluorescence polarization assay |
Eur J Med Chem 136: 14-35 (2017)
Article DOI: 10.1016/j.ejmech.2017.04.047
BindingDB Entry DOI: 10.7270/Q2F76G3G |
More data for this
Ligand-Target Pair | |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data