 Found 32 hits of kd data for polymerid = 50006167,50006362,50006490,50006781,50007060,50007089,5291,5691,8232
Found 32 hits of kd data for polymerid = 50006167,50006362,50006490,50006781,50007060,50007089,5291,5691,8232 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Alpha-ketoglutarate-dependent dioxygenase FTO
(Homo sapiens (Human)) | BDBM50596052
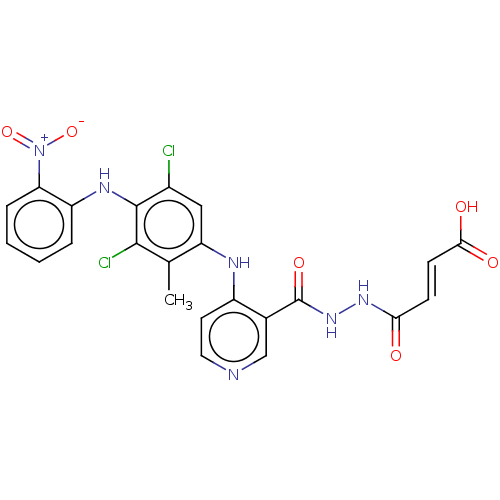
(CHEMBL5196484)Show SMILES Cc1c(Nc2ccncc2C(=O)NNC(=O)\C=C\C(O)=O)cc(Cl)c(Nc2ccccc2[N+]([O-])=O)c1Cl | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 28 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.1c01107
BindingDB Entry DOI: 10.7270/Q2QF8XWG |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 4D
(Homo sapiens (Human)) | BDBM50574750

(CHEMBL4850758)Show SMILES CC(C)(C)OC(=O)N[C@@H]1CCN(C1)C(c1ccc(cc1)-c1ccncc1)c1cc2OCOc2cc1O |r| | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 36 | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Inhibition of human KDM4D expressed in Escherichia coli BL21 (DE3) assessed as equilibrium dissociation constant measured by isothermal titration cal... |
Citation and Details
Article DOI: 10.1016/j.ejmech.2021.113662
BindingDB Entry DOI: 10.7270/Q24T6P5X |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245000
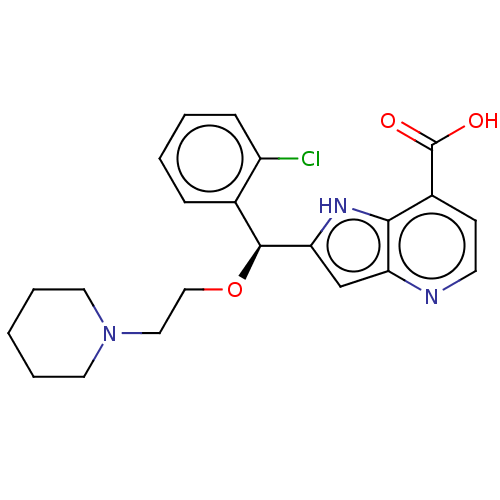
(CHEMBL4096760)Show SMILES OC(=O)c1ccnc2cc([nH]c12)[C@@H](OCCN1CCCCC1)c1ccccc1Cl |r| Show InChI InChI=1S/C22H24ClN3O3/c23-17-7-3-2-6-15(17)21(29-13-12-26-10-4-1-5-11-26)19-14-18-20(25-19)16(22(27)28)8-9-24-18/h2-3,6-9,14,21,25H,1,4-5,10-13H2,(H,27,28)/t21-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 60 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245002
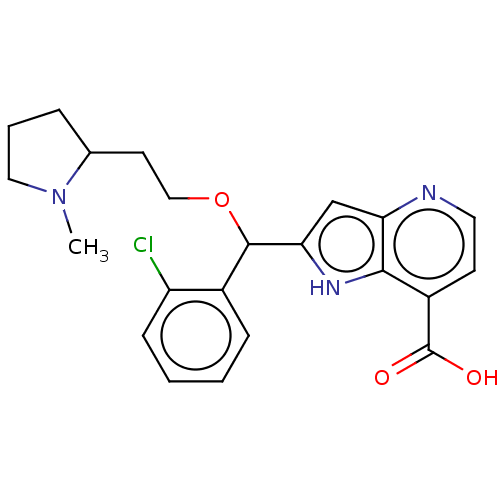
(CHEMBL4081571)Show SMILES CN1CCCC1CCOC(c1cc2nccc(C(O)=O)c2[nH]1)c1ccccc1Cl Show InChI InChI=1S/C22H24ClN3O3/c1-26-11-4-5-14(26)9-12-29-21(15-6-2-3-7-17(15)23)19-13-18-20(25-19)16(22(27)28)8-10-24-18/h2-3,6-8,10,13-14,21,25H,4-5,9,11-12H2,1H3,(H,27,28) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 150 | n/a | n/a | n/a | n/a | n/a |
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assay |
J Med Chem 61: 10588-10601 (2018)
Article DOI: 10.1021/acs.jmedchem.8b01219
BindingDB Entry DOI: 10.7270/Q2R49TGF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245002

(CHEMBL4081571)Show SMILES CN1CCCC1CCOC(c1cc2nccc(C(O)=O)c2[nH]1)c1ccccc1Cl Show InChI InChI=1S/C22H24ClN3O3/c1-26-11-4-5-14(26)9-12-29-21(15-6-2-3-7-17(15)23)19-13-18-20(25-19)16(22(27)28)8-10-24-18/h2-3,6-8,10,13-14,21,25H,4-5,9,11-12H2,1H3,(H,27,28) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 150 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity at [3H]QNB and GppNHp radiolabeled muscarinic M2 receptor in rat heart. |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245004

(CHEMBL4090351)Show SMILES OC(=O)c1ccnc2cc([nH]c12)C(OCCN1CCCCC1)c1ccccc1Cl Show InChI InChI=1S/C22H24ClN3O3/c23-17-7-3-2-6-15(17)21(29-13-12-26-10-4-1-5-11-26)19-14-18-20(25-19)16(22(27)28)8-9-24-18/h2-3,6-9,14,21,25H,1,4-5,10-13H2,(H,27,28) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 180 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245005

(CHEMBL4070992)Show SMILES OC(=O)c1ccnc2cc([nH]c12)C(OCCN1CCC(F)(F)CC1)c1ccccc1Cl Show InChI InChI=1S/C22H22ClF2N3O3/c23-16-4-2-1-3-14(16)20(31-12-11-28-9-6-22(24,25)7-10-28)18-13-17-19(27-18)15(21(29)30)5-8-26-17/h1-5,8,13,20,27H,6-7,9-12H2,(H,29,30) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 210 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245037
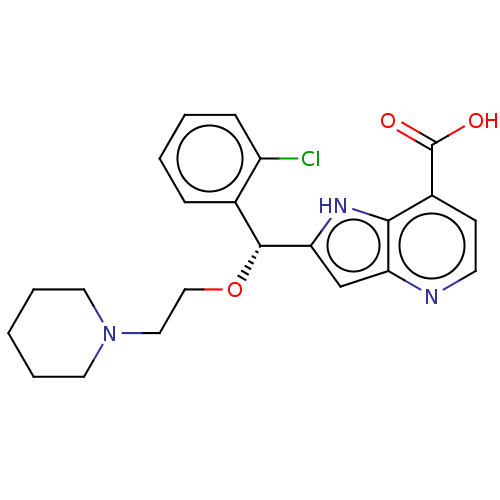
(CHEMBL4062359)Show SMILES OC(=O)c1ccnc2cc([nH]c12)[C@H](OCCN1CCCCC1)c1ccccc1Cl |r| Show InChI InChI=1S/C22H24ClN3O3/c23-17-7-3-2-6-15(17)21(29-13-12-26-10-4-1-5-11-26)19-14-18-20(25-19)16(22(27)28)8-9-24-18/h2-3,6-9,14,21,25H,1,4-5,10-13H2,(H,27,28)/t21-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 220 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity at [3H]QNB radiolabeled muscarinic M2 receptor in rat heart. |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245003
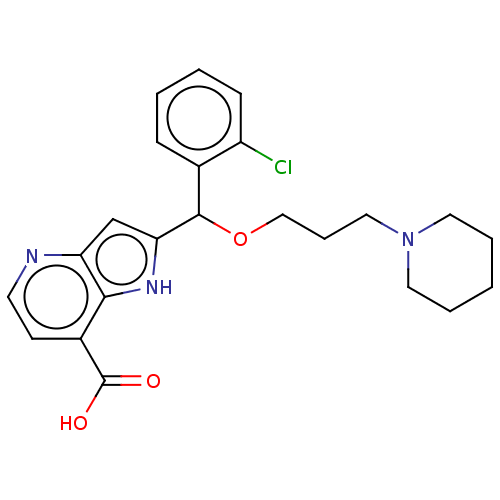
(CHEMBL4066179)Show SMILES OC(=O)c1ccnc2cc([nH]c12)C(OCCCN1CCCCC1)c1ccccc1Cl Show InChI InChI=1S/C23H26ClN3O3/c24-18-8-3-2-7-16(18)22(30-14-6-13-27-11-4-1-5-12-27)20-15-19-21(26-20)17(23(28)29)9-10-25-19/h2-3,7-10,15,22,26H,1,4-6,11-14H2,(H,28,29) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 250 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50468054
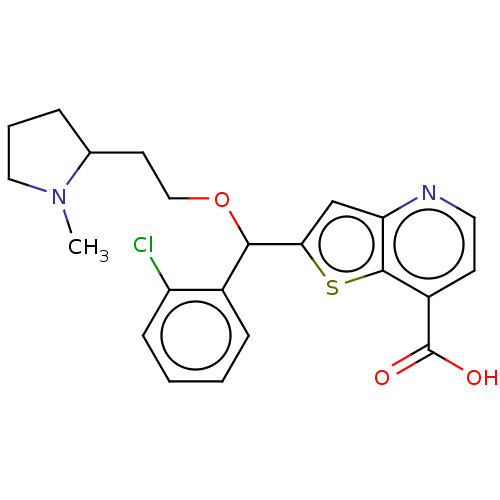
(CHEMBL4284220)Show SMILES CN1CCCC1CCOC(c1cc2nccc(C(O)=O)c2s1)c1ccccc1Cl Show InChI InChI=1S/C22H23ClN2O3S/c1-25-11-4-5-14(25)9-12-28-20(15-6-2-3-7-17(15)23)19-13-18-21(29-19)16(22(26)27)8-10-24-18/h2-3,6-8,10,13-14,20H,4-5,9,11-12H2,1H3,(H,26,27) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 260 | n/a | n/a | n/a | n/a | n/a |
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assay |
J Med Chem 61: 10588-10601 (2018)
Article DOI: 10.1021/acs.jmedchem.8b01219
BindingDB Entry DOI: 10.7270/Q2R49TGF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Alpha-ketoglutarate-dependent dioxygenase FTO
(Homo sapiens (Human)) | BDBM50606681

(CHEMBL5219846)Show SMILES OC(=O)CNC(=O)c1nc(Nc2ccc(cc2)[N+]([O-])=O)ccc1O | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 340 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.1c01204
BindingDB Entry DOI: 10.7270/Q2BR8X83 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245006
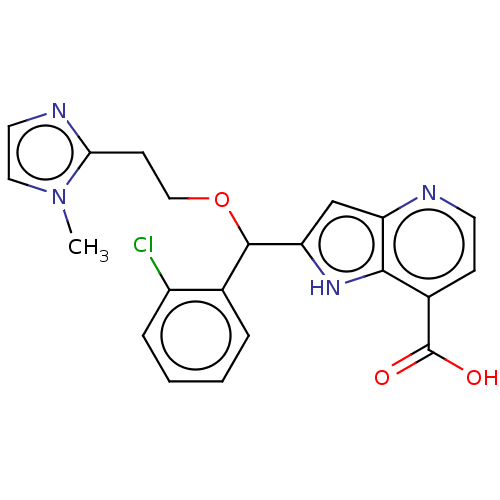
(CHEMBL4077310)Show SMILES Cn1ccnc1CCOC(c1cc2nccc(C(O)=O)c2[nH]1)c1ccccc1Cl Show InChI InChI=1S/C21H19ClN4O3/c1-26-10-9-24-18(26)7-11-29-20(13-4-2-3-5-15(13)22)17-12-16-19(25-17)14(21(27)28)6-8-23-16/h2-6,8-10,12,20,25H,7,11H2,1H3,(H,27,28) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 410 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50245001

(CHEMBL4059617)Show SMILES OC(=O)c1ccnc2cc([nH]c12)C(OCC1CCC(F)(F)CC1)c1ccccc1Cl Show InChI InChI=1S/C22H21ClF2N2O3/c23-16-4-2-1-3-14(16)20(30-12-13-5-8-22(24,25)9-6-13)18-11-17-19(27-18)15(21(28)29)7-10-26-17/h1-4,7,10-11,13,20,27H,5-6,8-9,12H2,(H,28,29) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 470 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM320423

(2-[(2-chlorophenyl)-propoxy- methyl]-1H-pyrrolo[3,...) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 530 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Alpha-ketoglutarate-dependent dioxygenase FTO
(Homo sapiens (Human)) | BDBM50606686

(CHEMBL5219336)Show SMILES CNC(=O)c1ccc(Nc2ccc(O)c(n2)C(=O)NCC(O)=O)cc1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 670 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.1c01204
BindingDB Entry DOI: 10.7270/Q2BR8X83 |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50282115
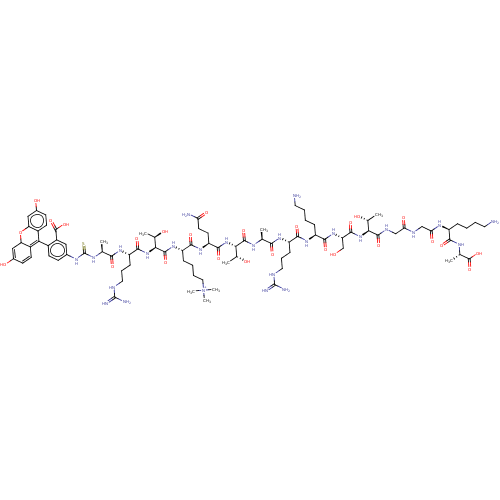
(CHEMBL4165111)Show SMILES C[C@@H](O)[C@H](NC(=O)[C@H](CO)NC(=O)[C@H](CCCCN)NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=O)[C@@H](NC(=O)[C@H](CCC(N)=O)NC(=O)[C@H](CCCC[N+](C)(C)C)NC(=O)[C@@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=S)Nc1ccc(C2=C3C=CC(O)=CC3Oc3cc(O)ccc23)c(c1)C(O)=O)[C@@H](C)O)[C@@H](C)O)C(=O)NCC(=O)NCC(=O)N[C@@H](CCCCN)C(=O)N[C@@H](C)C(O)=O |r,c:88,90,93| Show InChI InChI=1S/C87H136N26O26S/c1-42(71(123)104-58(21-16-33-95-85(91)92)75(127)106-56(19-11-14-32-89)76(128)109-61(41-114)79(131)110-68(45(4)115)80(132)98-39-65(121)97-40-66(122)103-55(18-10-13-31-88)73(125)100-44(3)83(135)136)99-81(133)69(46(5)116)111-78(130)60(29-30-64(90)120)107-74(126)57(20-12-15-35-113(7,8)9)108-82(134)70(47(6)117)112-77(129)59(22-17-34-96-86(93)94)105-72(124)43(2)101-87(140)102-48-23-26-51(54(36-48)84(137)138)67-52-27-24-49(118)37-62(52)139-63-38-50(119)25-28-53(63)67/h23-28,36-38,42-47,55-62,68-70,114-117H,10-22,29-35,39-41,88-89H2,1-9H3,(H29-,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,140)/p+1/t42-,43-,44-,45+,46+,47+,55-,56-,57-,58-,59-,60-,61-,62?,68-,69-,70-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| KEGG
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | n/a | 800 | n/a | n/a | n/a | n/a | n/a |
University of Connecticut
Curated by ChEMBL
| Assay Description
Inhibition of JARID1A (unknown origin) |
Eur J Med Chem 136: 14-35 (2017)
Article DOI: 10.1016/j.ejmech.2017.04.047
BindingDB Entry DOI: 10.7270/Q2F76G3G |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50282162

(CHEMBL4173016)Show SMILES C[C@@H](O)[C@H](NC(=O)[C@H](CO)NC(=O)[C@H](CCCCN)NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=O)[C@@H](NC(=O)[C@H](CCC(N)=O)NC(=O)[C@H](CCCCN(C)C)NC(=O)[C@@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@H](C)NC(=S)Nc1ccc(C2=C3C=CC(O)=CC3Oc3cc(O)ccc23)c(c1)C(O)=O)[C@@H](C)O)[C@@H](C)O)C(=O)NCC(=O)NCC(=O)N[C@@H](CCCCN)C(=O)N[C@@H](C)C(O)=O |r,c:87,89,92| Show InChI InChI=1S/C86H134N26O26S/c1-41(70(122)103-57(20-15-32-94-84(90)91)74(126)105-55(18-10-13-31-88)75(127)108-60(40-113)78(130)109-67(44(4)114)79(131)97-38-64(120)96-39-65(121)102-54(17-9-12-30-87)72(124)99-43(3)82(134)135)98-80(132)68(45(5)115)110-77(129)59(28-29-63(89)119)106-73(125)56(19-11-14-34-112(7)8)107-81(133)69(46(6)116)111-76(128)58(21-16-33-95-85(92)93)104-71(123)42(2)100-86(139)101-47-22-25-50(53(35-47)83(136)137)66-51-26-23-48(117)36-61(51)138-62-37-49(118)24-27-52(62)66/h22-27,35-37,41-46,54-61,67-69,113-118H,9-21,28-34,38-40,87-88H2,1-8H3,(H2,89,119)(H,96,120)(H,97,131)(H,98,132)(H,99,124)(H,102,121)(H,103,122)(H,104,123)(H,105,126)(H,106,125)(H,107,133)(H,108,127)(H,109,130)(H,110,129)(H,111,128)(H,134,135)(H,136,137)(H4,90,91,94)(H4,92,93,95)(H2,100,101,139)/t41-,42-,43-,44+,45+,46+,54-,55-,56-,57-,58-,59-,60-,61?,67-,68-,69-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| KEGG
PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | n/a | 800 | n/a | n/a | n/a | n/a | n/a |
University of Connecticut
Curated by ChEMBL
| Assay Description
Inhibition of JARID1A (unknown origin) |
Eur J Med Chem 136: 14-35 (2017)
Article DOI: 10.1016/j.ejmech.2017.04.047
BindingDB Entry DOI: 10.7270/Q2F76G3G |
More data for this
Ligand-Target Pair | |
Alpha-ketoglutarate-dependent dioxygenase FTO
(Homo sapiens (Human)) | BDBM50606679
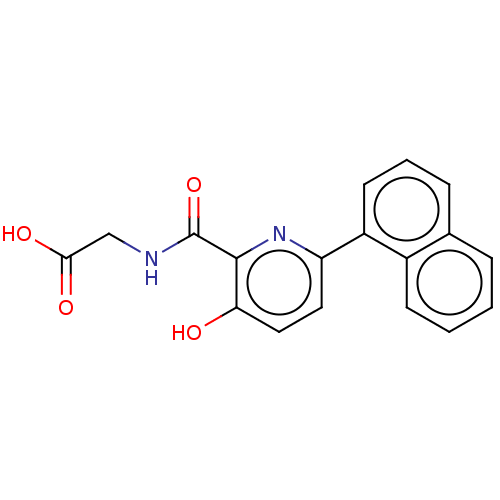
(CHEMBL5218636) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 1.20E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.1c01204
BindingDB Entry DOI: 10.7270/Q2BR8X83 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 6A
(Homo sapiens (Human)) | BDBM231631
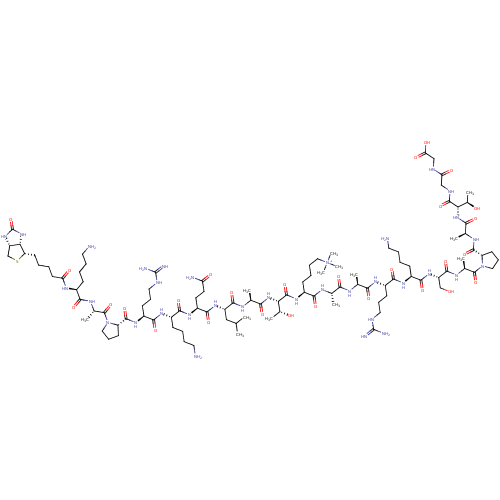
(Biotin-KAPRKQLATKAARKMe3SAPATGG | H3K27Me3 peptide)Show SMILES CC(C)C[C@H](NC(=O)[C@H](CCC(N)=O)NC(=O)[C@H](CCCCN)NC(=O)[C@H](CCCNC(N)=N)NC(=O)[C@@H]1CCCN1C(=O)[C@H](C)NC(=O)[C@H](CCCCN)NC(=O)CCCC[C@@H]1SC[C@@H]2NC(=O)N[C@H]12)C(=O)N[C@@H](C)C(=O)N[C@@H]([C@@H](C)O)C(=O)N[C@@H](CCCC[N+](C)(C)C)C(=O)N[C@@H](C)C(=O)N[C@@H](C)C(=O)N[C@@H](CCCNC(N)=N)C(=O)N[C@@H](CCCCN)C(=O)N[C@@H](CO)C(=O)N[C@@H](C)C(=O)N1CCC[C@H]1C(=O)N[C@@H](C)C(=O)N[C@@H]([C@@H](C)O)C(=O)NCC(=O)NCC(O)=O |r| Show InChI InChI=1S/C103H182N34O28S/c1-53(2)48-69(129-92(156)68(38-39-75(107)141)126-88(152)64(29-17-21-41-105)124-90(154)67(33-25-44-113-102(110)111)127-96(160)73-35-27-46-136(73)99(163)58(7)120-86(150)62(28-16-20-40-104)122-76(142)37-15-14-36-74-81-71(52-166-74)131-103(165)134-81)93(157)118-56(5)84(148)133-80(61(10)140)98(162)128-63(31-19-23-47-137(11,12)13)87(151)117-54(3)82(146)116-55(4)83(147)123-66(32-24-43-112-101(108)109)89(153)125-65(30-18-22-42-106)91(155)130-70(51-138)94(158)121-59(8)100(164)135-45-26-34-72(135)95(159)119-57(6)85(149)132-79(60(9)139)97(161)115-49-77(143)114-50-78(144)145/h53-74,79-81,138-140H,14-52,104-106H2,1-13H3,(H31-,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,165)/p+1/t54-,55-,56-,57-,58-,59-,60+,61+,62-,63-,64-,65-,66-,67-,68-,69-,70-,71-,72-,73-,74-,79-,80-,81-/m0/s1 | PDB
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| KEGG
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 2.00E+3 | n/a | n/a | n/a | 7.5 | 25 |
University of Oxford
| Assay Description
Binding assays were carried out as 20-μl reactions in 384-well white ProxiPlates (Perkin-Elmer Life Sciences) as described [Hong et al., Proc. N... |
J Biol Chem 289: 18302-13 (2014)
Article DOI: 10.1074/jbc.M114.555052
BindingDB Entry DOI: 10.7270/Q23X85J4 |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM281089
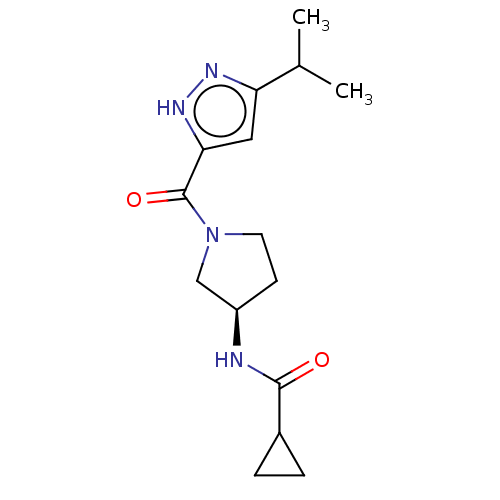
(US10022354, Example 29)Show SMILES CC(C)c1cc([nH]n1)C(=O)N1CC[C@H](C1)NC(=O)C1CC1 |r| Show InChI InChI=1S/C15H22N4O2/c1-9(2)12-7-13(18-17-12)15(21)19-6-5-11(8-19)16-14(20)10-3-4-10/h7,9-11H,3-6,8H2,1-2H3,(H,16,20)(H,17,18)/t11-/m1/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 2.40E+3 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Probable JmjC domain-containing histone demethylation protein 2C
(Homo sapiens (Human)) | BDBM50594964
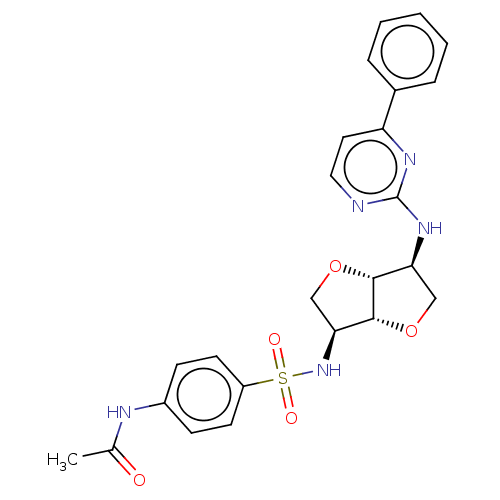
(CHEMBL5193187)Show SMILES [H][C@]12OC[C@H](NS(=O)(=O)c3ccc(NC(C)=O)cc3)[C@@]1([H])OC[C@@H]2Nc1nccc(n1)-c1ccccc1 |r| | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 2.70E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1016/j.ejmech.2022.114143
BindingDB Entry DOI: 10.7270/Q28W3J9V |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM223320
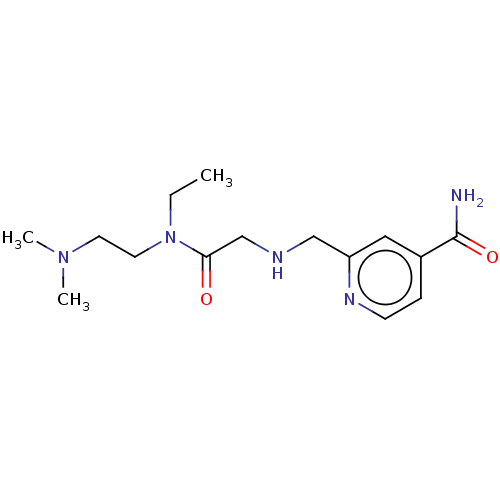
(KDOAM-25)Show InChI InChI=1S/C15H25N5O2/c1-4-20(8-7-19(2)3)14(21)11-17-10-13-9-12(15(16)22)5-6-18-13/h5-6,9,17H,4,7-8,10-11H2,1-3H3,(H2,16,22) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 2.70E+3 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Alpha-ketoglutarate-dependent dioxygenase FTO
(Homo sapiens (Human)) | BDBM50606687

(CHEMBL5220246)Show SMILES CN(C)C(=O)c1ccc(Nc2ccc(O)c(n2)C(=O)NCC(O)=O)cc1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 3.90E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1021/acs.jmedchem.1c01204
BindingDB Entry DOI: 10.7270/Q2BR8X83 |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50468052
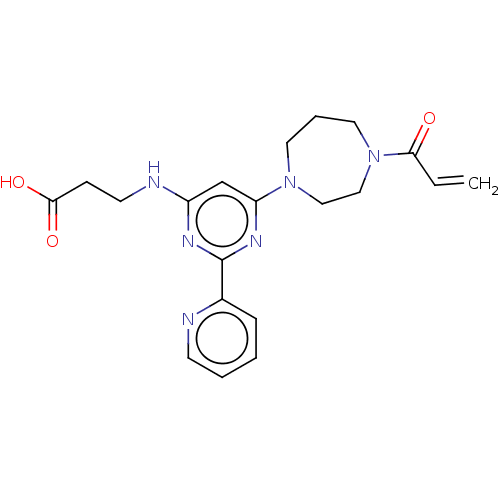
(CHEMBL4283844)Show SMILES OC(=O)CCNc1cc(nc(n1)-c1ccccn1)N1CCCN(CC1)C(=O)C=C Show InChI InChI=1S/C20H24N6O3/c1-2-18(27)26-11-5-10-25(12-13-26)17-14-16(22-9-7-19(28)29)23-20(24-17)15-6-3-4-8-21-15/h2-4,6,8,14H,1,5,7,9-13H2,(H,28,29)(H,22,23,24) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 6.00E+3 | n/a | n/a | n/a | n/a | n/a |
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assay |
J Med Chem 61: 10588-10601 (2018)
Article DOI: 10.1021/acs.jmedchem.8b01219
BindingDB Entry DOI: 10.7270/Q2R49TGF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Alpha-ketoglutarate-dependent dioxygenase alkB homolog 3
(Homo sapiens (Human)) | BDBM50463170
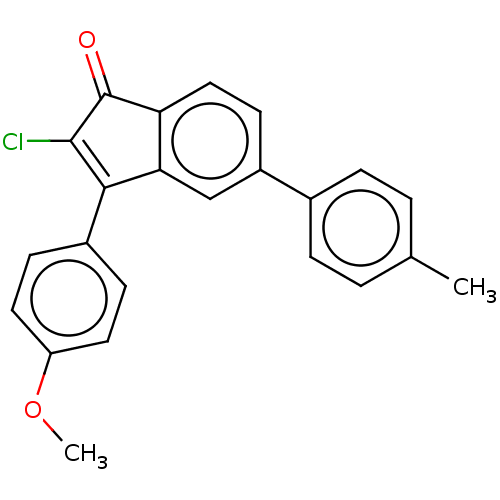
(CHEMBL4239759)Show SMILES COc1ccc(cc1)C1=C(Cl)C(=O)c2ccc(cc12)-c1ccc(C)cc1 |c:9| Show InChI InChI=1S/C23H17ClO2/c1-14-3-5-15(6-4-14)17-9-12-19-20(13-17)21(22(24)23(19)25)16-7-10-18(26-2)11-8-16/h3-13H,1-2H3 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 7.81E+3 | n/a | n/a | n/a | n/a | n/a |
Indian Institute of Technology Hyderabad
Curated by ChEMBL
| Assay Description
Binding affinity to recombinant human AlkBH3 incubated for 2 mins by isothermal titration calorimetric method |
Bioorg Med Chem 26: 4100-4112 (2018)
Article DOI: 10.1016/j.bmc.2018.06.040
BindingDB Entry DOI: 10.7270/Q22B91PG |
More data for this
Ligand-Target Pair | |
Probable JmjC domain-containing histone demethylation protein 2C
(Homo sapiens (Human)) | BDBM50594965
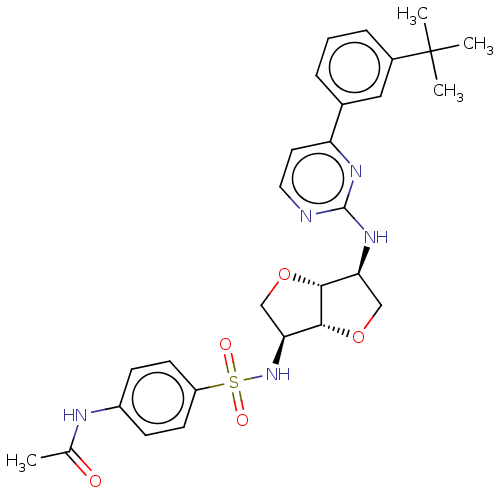
(CHEMBL5173150)Show SMILES [H][C@]12OC[C@H](NS(=O)(=O)c3ccc(NC(C)=O)cc3)[C@@]1([H])OC[C@@H]2Nc1nccc(n1)-c1cccc(c1)C(C)(C)C |r| | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 8.06E+3 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1016/j.ejmech.2022.114143
BindingDB Entry DOI: 10.7270/Q28W3J9V |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM50468053

(CHEMBL4294426)Show SMILES OC(=O)CCNc1cc(nc(n1)-c1ccccn1)N1CCCN(CC1)S(=O)(=O)C=C Show InChI InChI=1S/C19H24N6O4S/c1-2-30(28,29)25-11-5-10-24(12-13-25)17-14-16(21-9-7-18(26)27)22-19(23-17)15-6-3-4-8-20-15/h2-4,6,8,14H,1,5,7,9-13H2,(H,26,27)(H,21,22,23) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 9.00E+3 | n/a | n/a | n/a | n/a | n/a |
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assay |
J Med Chem 61: 10588-10601 (2018)
Article DOI: 10.1021/acs.jmedchem.8b01219
BindingDB Entry DOI: 10.7270/Q2R49TGF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM281088
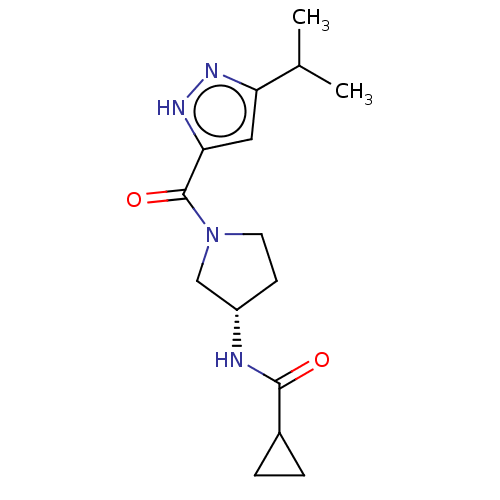
(US10022354, Example 28)Show SMILES CC(C)c1cc([nH]n1)C(=O)N1CC[C@@H](C1)NC(=O)C1CC1 |r| Show InChI InChI=1S/C15H22N4O2/c1-9(2)12-7-13(18-17-12)15(21)19-6-5-11(8-19)16-14(20)10-3-4-10/h7,9-11H,3-6,8H2,1-2H3,(H,16,20)(H,17,18)/t11-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | >1.20E+4 | n/a | n/a | n/a | n/a | n/a |
The University of Texas MD Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to ARID/PHD1/Zn binding domain truncated human N-terminal His-SUMO-tagged KDM5A (1 to 588 residues) expressed in Escherichia coli by... |
J Med Chem 61: 3193-3208 (2018)
Article DOI: 10.1021/acs.jmedchem.8b00261
BindingDB Entry DOI: 10.7270/Q2BC41Z0 |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 5A
(Homo sapiens (Human)) | BDBM60875
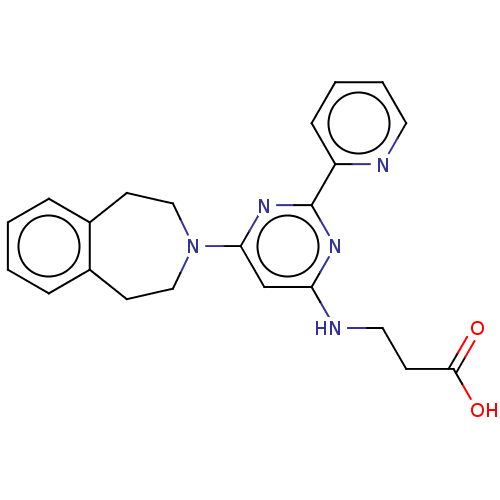
(3-((6-(4,5-Dihydro-1H-benzo[d]azepin-3(2H)-yl)-2-(...)Show SMILES OC(=O)CCNc1cc(nc(n1)-c1ccccn1)N1CCc2ccccc2CC1 Show InChI InChI=1S/C22H23N5O2/c28-21(29)8-12-24-19-15-20(26-22(25-19)18-7-3-4-11-23-18)27-13-9-16-5-1-2-6-17(16)10-14-27/h1-7,11,15H,8-10,12-14H2,(H,28,29)(H,24,25,26) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
MCE
PC cid
PC sid
PDB
UniChem
| PDB
Article
PubMed
| n/a | n/a | n/a | 1.20E+4 | n/a | n/a | n/a | n/a | n/a |
The University of Texas M.D. Anderson Cancer Center
Curated by ChEMBL
| Assay Description
Binding affinity to KDM5A ARID/PhD1 domain deletion mutant (1 to 588 residues) (unknown origin) by ITC assay |
J Med Chem 61: 10588-10601 (2018)
Article DOI: 10.1021/acs.jmedchem.8b01219
BindingDB Entry DOI: 10.7270/Q2R49TGF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Alpha-ketoglutarate-dependent dioxygenase FTO
(Homo sapiens (Human)) | BDBM50117830
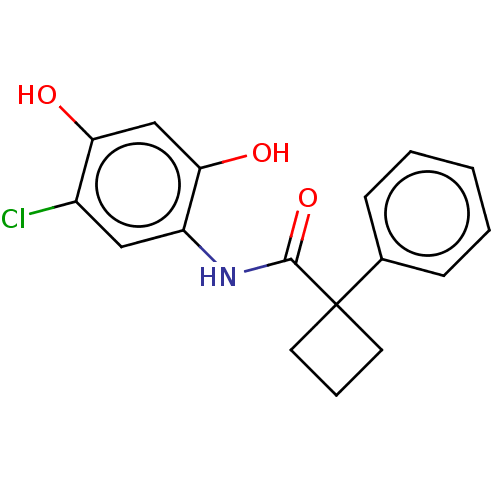
(CHEMBL3613953)Show InChI InChI=1S/C17H16ClNO3/c18-12-9-13(15(21)10-14(12)20)19-16(22)17(7-4-8-17)11-5-2-1-3-6-11/h1-3,5-6,9-10,20-21H,4,7-8H2,(H,19,22) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
PC cid
PC sid
PDB
UniChem
Similars
| PDB
Article
PubMed
| n/a | n/a | n/a | 1.30E+4 | n/a | n/a | n/a | n/a | n/a |
Zhengzhou University
Curated by ChEMBL
| Assay Description
Binding affinity to FTO (unknown origin) (31 to 505 residues) expressed in Escherichia coli BL21 (DE3) by ITC method |
J Med Chem 58: 7341-8 (2015)
Article DOI: 10.1021/acs.jmedchem.5b00702
BindingDB Entry DOI: 10.7270/Q2X068TB |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Probable JmjC domain-containing histone demethylation protein 2C
(Homo sapiens (Human)) | BDBM50594963
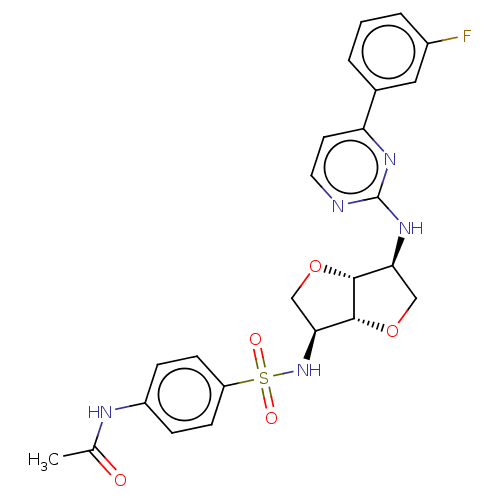
(CHEMBL5191875)Show SMILES [H][C@]12OC[C@H](NS(=O)(=O)c3ccc(NC(C)=O)cc3)[C@@]1([H])OC[C@@H]2Nc1nccc(n1)-c1cccc(F)c1 |r| | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 2.67E+4 | n/a | n/a | n/a | n/a | n/a |
TBA
| |
Citation and Details
Article DOI: 10.1016/j.ejmech.2022.114143
BindingDB Entry DOI: 10.7270/Q28W3J9V |
More data for this
Ligand-Target Pair | |
Lysine-specific demethylase 4B
(Homo sapiens (Human)) | BDBM50457513

(CHEMBL4212908)Show InChI InChI=1S/C16H23NO/c1-17(2)11-14-12-8-9-16(10-12,15(14)18)13-6-4-3-5-7-13/h3-7,12,14-15,18H,8-11H2,1-2H3/t12?,14-,15-,16?/m0/s1 | PDB
UniProtKB/SwissProt
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 1.27E+5 | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc.
Curated by ChEMBL
| Assay Description
Binding affinity to 6His-tagged and human KDM4B tandem TUDOR domain (916 to 1030 residues) expressed in Escherichia coli BL21(DE3)-T1R by ITC method |
Bioorg Med Chem Lett 28: 1708-1713 (2018)
Article DOI: 10.1016/j.bmcl.2018.04.050
BindingDB Entry DOI: 10.7270/Q2WM1H1X |
More data for this
Ligand-Target Pair | |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data