

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Iodotyrosine deiodinase 1 [1-1,32-289] (Homo sapiens (Human)) | BDBM37633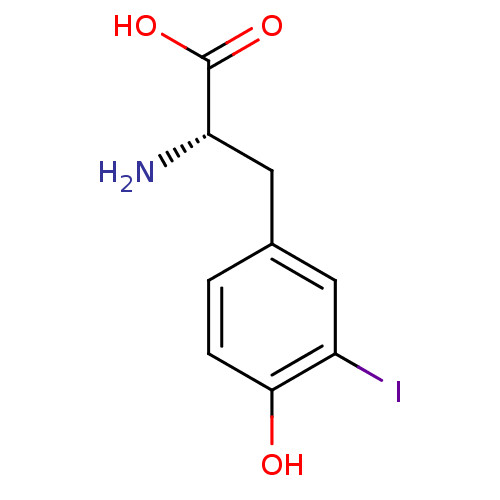 ((2S)-2-amino-3-(4-hydroxy-3-iodo-phenyl)propionic ...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | n/a | 90 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Iodotyrosine deiodinase [1-228] (Haliscomenobacter hydrossis) | BDBM217400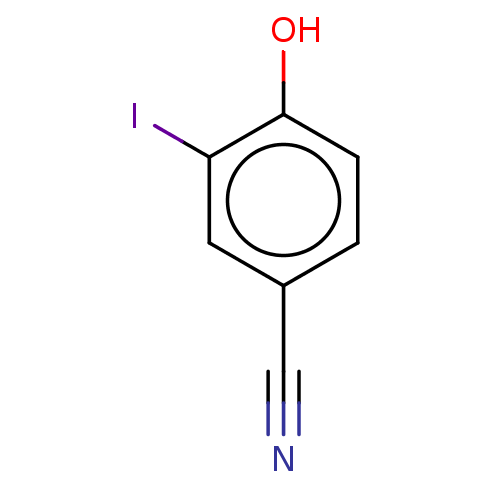 (4-Cyano-2-iodophenol (2IPCN)) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.44E+3 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Iodotyrosine deiodinase [1-228] (Haliscomenobacter hydrossis) | BDBM37633 ((2S)-2-amino-3-(4-hydroxy-3-iodo-phenyl)propionic ...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | n/a | 8.20E+3 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Iodotyrosine deiodinase [1-228] (Haliscomenobacter hydrossis) | BDBM217399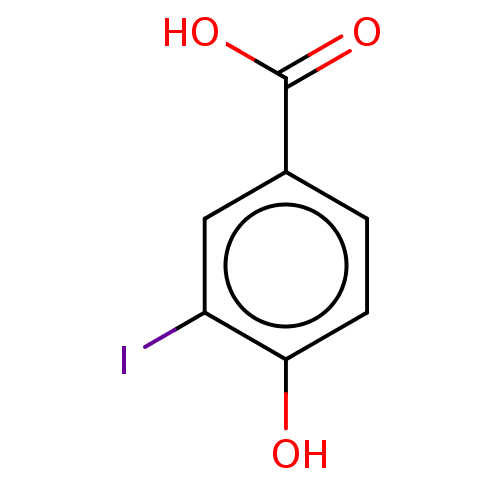 (4-Hydroxy-3-iodobenzoic acid (2IPCOOH)) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | n/a | 2.41E+4 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Iodotyrosine deiodinase [1-228] (Haliscomenobacter hydrossis) | BDBM217397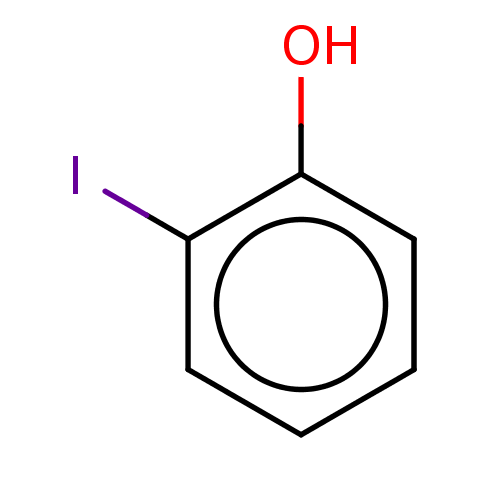 (2-Iodophenol (2IP)) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE KEGG PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 6.70E+4 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Iodotyrosine deiodinase [1-228] (Haliscomenobacter hydrossis) | BDBM217398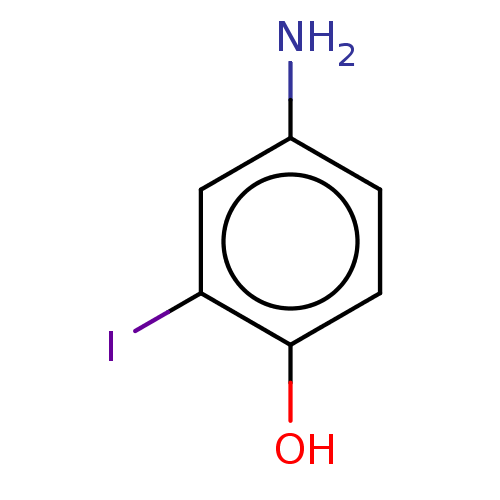 (4-Amino-2-iodophenol (4A2IP)) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 9.60E+4 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Iodotyrosine deiodinase 1 [1-1,32-289] (Homo sapiens (Human)) | BDBM217397 (2-Iodophenol (2IP)) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE KEGG PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | n/a | 1.41E+6 | n/a | n/a | n/a | n/a | n/a |
Johns Hopkins University | Assay Description Dissociation constants (Kd) for the ligands of IYD were determined by their ability to quench the fluorescence of the oxidized FMN (FMNox) (λex ... | Biochemistry 56: 1130-1139 (2017) Article DOI: 10.1021/acs.biochem.6b01308 BindingDB Entry DOI: 10.7270/Q2ZK5FJM | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||