

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coagulation factor X (94/94 = 100%)† (Homo sapiens (Human)) | BDBM17295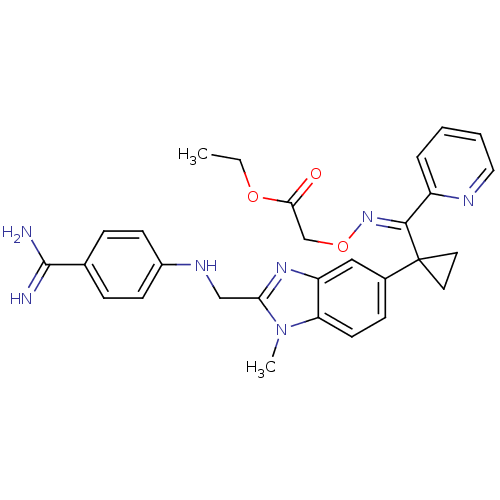 (BIBT0871 | ethyl 2-{[(E)-{[1-(2-{[(4-carbamimidoyl...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem | PDB Article PubMed | 57 | -43.0 | n/a | n/a | n/a | n/a | n/a | 8.0 | 37 |
Boehringer Ingelheim Pharma KG | Assay Description For determination of the inhibition constants (Ki), enzyme inhibition was studied at three different substrate concentrations and seven different con... | Structure 9: 29-37 (2001) Article DOI: 10.1016/s0969-2126(00)00551-7 BindingDB Entry DOI: 10.7270/Q2028PTC | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||