

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbonic anhydrase 2 [F130V] (258/259 > 99%)† (Homo sapiens (Human)) | BDBM12019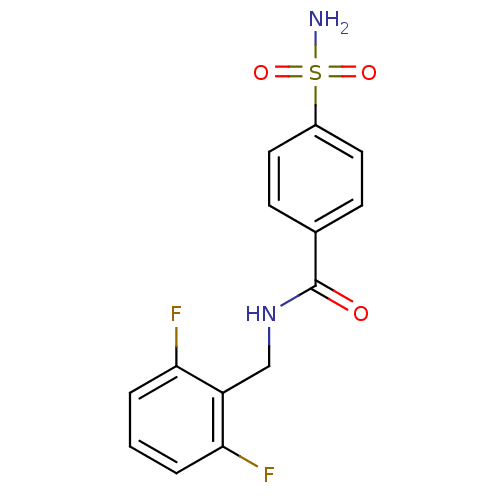 (2,6-difluoro-SBB | 4-(AMINOSULFONYL)-N-[(2,6-DIFLU...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 3.90 | n/a | n/a | n/a | 7.4 | 37 |
University of Pennsylvania | Assay Description Inhibitor binding to CA II was determined using a fluorescence competition assay. Displacement of dansylamide and binding of the inhibitor was determ... | J Am Chem Soc 123: 9620-7 (2001) Article DOI: 10.1021/ja011034p BindingDB Entry DOI: 10.7270/Q2QZ286J | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||