

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heat shock cognate 71 kDa protein (386/386 = 100%)† (Homo sapiens (Human)) | BDBM50013703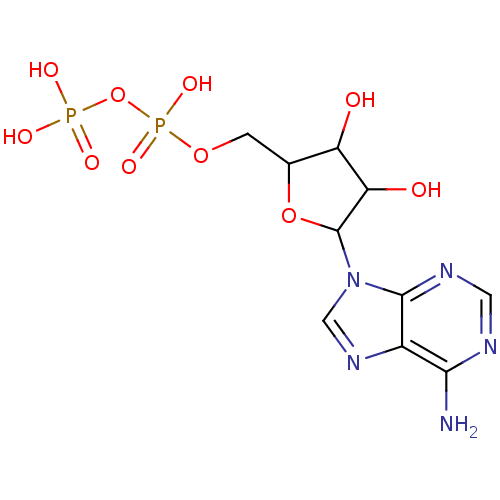 ((ADP)[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetra...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL KEGG MMDB PC cid PC sid PDB UniChem Similars | PDB PCBioAssay | n/a | n/a | 280 | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Center for Chemical Genomics Curated by PubChem BioAssay | Assay Description Data Source: Sanford-Burnham Center for Chemical Genomics (SBCCG) Source Affiliation: Sanford-Burnham Medical Research Institute (SBMRI, San Diego, C... | PubChem Bioassay (2008) BindingDB Entry DOI: 10.7270/Q20G3HKD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Heat shock cognate 71 kDa protein (386/386 = 100%)† (Homo sapiens (Human)) | BDBM50368125 (ADENOSINE DIPHOSPHATE | ADP) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 320 | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research Curated by ChEMBL | Assay Description Binding affinity to human truncated HSC70 NBD (1 to 381 residues) by SPR analysis | J Med Chem 59: 4625-36 (2016) Article DOI: 10.1021/acs.jmedchem.5b02001 BindingDB Entry DOI: 10.7270/Q2Z03B3X | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Heat shock cognate 71 kDa protein (386/386 = 100%)† (Homo sapiens (Human)) | BDBM50368125 (ADENOSINE DIPHOSPHATE | ADP) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 324 | n/a | n/a | n/a | n/a | n/a |
The Institute of Cancer Research Curated by ChEMBL | Assay Description Binding affinity to human truncated HSC70 NBD (1 to 381 residues) by SPR analysis | J Med Chem 59: 4625-36 (2016) Article DOI: 10.1021/acs.jmedchem.5b02001 BindingDB Entry DOI: 10.7270/Q2Z03B3X | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||