

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serine protease 1 (223/223 = 100%)† (Bos taurus (bovine)) | BDBM14153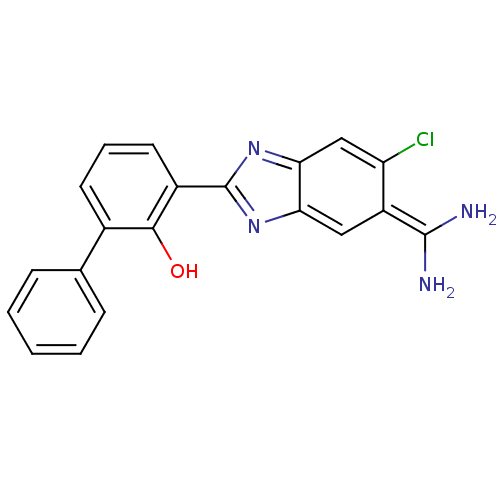 (2-{5-[amino(iminiumyl)methyl]-6-chloro-1H-1,3-benz...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 810 | -34.4 | n/a | n/a | n/a | n/a | n/a | 7.4 | 22 |
Celera | Assay Description Each enzyme was assayed with a set of different concentrations of each inhibitor. After addition of the appropriate substrate, the rate of hydrolysis... | J Mol Biol 329: 93-120 (2003) Article DOI: 10.1016/s0022-2836(03)00399-1 BindingDB Entry DOI: 10.7270/Q2R78CGQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine protease 1 (223/223 = 100%)† (Bos taurus (bovine)) | BDBM14153 (2-{5-[amino(iminiumyl)methyl]-6-chloro-1H-1,3-benz...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 2.10E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Axys Pharmaceutical | Assay Description Each enzyme was assayed with a set of different concentrations of each inhibitor. After addition of the appropriate substrate, the rate of hydrolysis... | Chem Biol 8: 1107-21 (2001) Article DOI: 10.1016/S1074-5521(01)00084-9 BindingDB Entry DOI: 10.7270/Q2S75DKN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine protease 1 (223/223 = 100%)† (Bos taurus (bovine)) | BDBM14153 (2-{5-[amino(iminiumyl)methyl]-6-chloro-1H-1,3-benz...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 3.30E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Axys Pharmaceutical | Assay Description Each enzyme was assayed with a set of different concentrations of each inhibitor. After addition of the appropriate substrate, the rate of hydrolysis... | Chem Biol 8: 1107-21 (2001) Article DOI: 10.1016/S1074-5521(01)00084-9 BindingDB Entry DOI: 10.7270/Q2S75DKN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine protease 1 (223/223 = 100%)† (Bos taurus (bovine)) | BDBM14153 (2-{5-[amino(iminiumyl)methyl]-6-chloro-1H-1,3-benz...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 4.20E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Axys Pharmaceutical | Assay Description Each enzyme was assayed with a set of different concentrations of each inhibitor. After addition of the appropriate substrate, the rate of hydrolysis... | Chem Biol 8: 1107-21 (2001) Article DOI: 10.1016/S1074-5521(01)00084-9 BindingDB Entry DOI: 10.7270/Q2S75DKN | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||