PDB code 1S9T
Identical Ligands in BindingDB
 Found 2 hits Enzyme Inhibition Constant Data
Found 2 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Glutamate receptor
(128/144 = 89%)†
(RABBIT) | BDBM17660
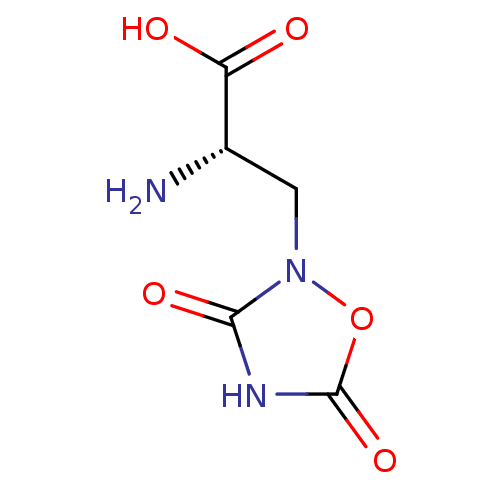
((2S)-2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl...)Show InChI InChI=1S/C5H7N3O5/c6-2(3(9)10)1-8-4(11)7-5(12)13-8/h2H,1,6H2,(H,9,10)(H,7,11,12)/t2-/m0/s1 | PDB
UniProtKB/TrEMBL
GoogleScholar
AffyNet 
| Purchase
CHEMBL
DrugBank
MCE
KEGG
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| 16.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Bristol
Curated by PDSP Ki Database
| |
Neuropharmacology 36: 1483-8 (1997)
Article DOI: 10.1016/s0028-3908(97)00161-5
BindingDB Entry DOI: 10.7270/Q2C827T6 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Glutamate receptor ionotropic, kainate 2
(141/144 = 98%)†
(Rattus norvegicus) | BDBM17660

((2S)-2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl...)Show InChI InChI=1S/C5H7N3O5/c6-2(3(9)10)1-8-4(11)7-5(12)13-8/h2H,1,6H2,(H,9,10)(H,7,11,12)/t2-/m0/s1 | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| Purchase
CHEMBL
DrugBank
MCE
KEGG
MMDB
PC cid
PC sid
PDB
UniChem
Patents
Similars
| DrugBank
PDB
Article
PubMed
| 134 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universite Blaise Pascal
Curated by ChEMBL
| Assay Description
Displacement of [3H]SYM2081 from rat recombinant iGluR6 |
J Med Chem 51: 4093-103 (2008)
Article DOI: 10.1021/jm800092x
BindingDB Entry DOI: 10.7270/Q20V8DP6 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
- Reorder this table by ec50
- Reorder this table by ic50
- Reorder this table by Kd
- Reorder this table by pH
- Reorder this table by T
- Reorder this table by ΔH°
- Reorder this table by -TΔS°
- Change energy units to: kcal/mole
Search BindingMOAD for More Affinity Data:
* indicates data uncertainty>20%
* 0.9 Tanimoto similarity
† Identities from BLAST output



 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data