

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lanosterol 14-alpha demethylase (449/451 > 99%)† (Mycobacterium tuberculosis (strain CDC 1551 / Oshk...) | BDBM25806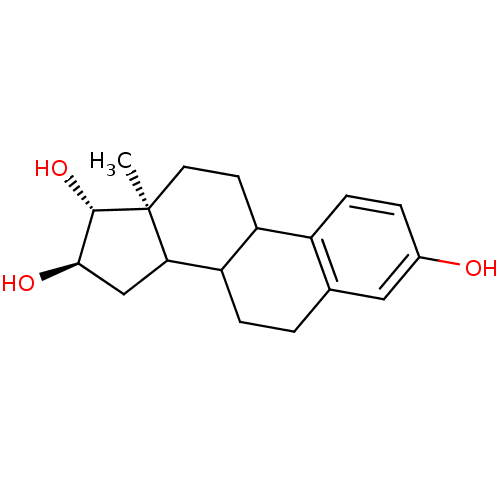 ((13R,14R,15S)-15-methyltetracyclo[8.7.0.0^{2,7}.0^...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | n/a | 1.00E+5 | n/a | n/a | n/a | 7.5 | 37 |
University of California at San Francisco | Assay Description The assay was developed based on the optical spectral property of P450 enzyme to elicit both type I and type II spectral changes (350 nm through 450 ... | Antimicrob Agents Chemother 51: 3915-23 (2007) Article DOI: 10.1128/AAC.00311-07 BindingDB Entry DOI: 10.7270/Q20V8B3R | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||