

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (231/232 > 99%)† (Escherichia coli) | BDBM22113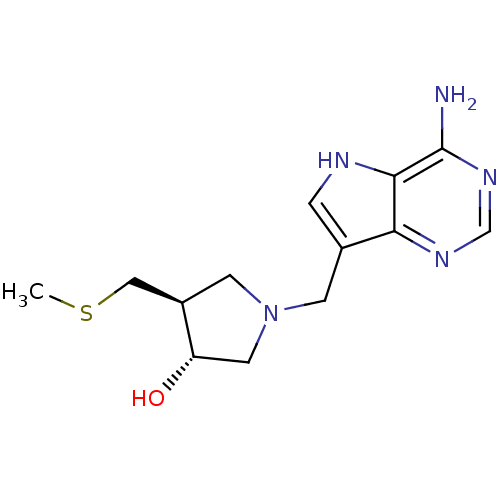 ((3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-y...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Victoria University of Wellington Curated by ChEMBL | Assay Description Inhibition of recombinant Escherichia coli MTAN expressed in Escherichia coli BL-21 DE3 using methylthioadenosine as substrate assessed as inhibition... | Bioorg Med Chem 23: 5326-33 (2015) Article DOI: 10.1016/j.bmc.2015.07.059 BindingDB Entry DOI: 10.7270/Q2HX1FF5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (232/232 = 100%)† (Escherichia coli (strain K12)) | BDBM22113 ((3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-y...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.0480 | -58.3 | n/a | n/a | n/a | n/a | n/a | 7.0 | 22 |
Industrial Research Limited | Assay Description Enzyme activity was monitored by absorbance change in the xanthine oxidase coupled assay, which measures the formation of 2,8-dihydroxyadenine at 293... | J Med Chem 51: 948-56 (2008) Article DOI: 10.1021/jm701265n BindingDB Entry DOI: 10.7270/Q2QC01T3 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (231/232 > 99%)† (Escherichia coli) | BDBM22113 ((3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-y...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.0480 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Victoria University of Wellington Curated by ChEMBL | Assay Description Inhibition of recombinant Escherichia coli MTAN expressed in Escherichia coli BL-21 DE3 using methylthioadenosine as substrate by xanthine oxidase co... | Bioorg Med Chem 23: 5326-33 (2015) Article DOI: 10.1016/j.bmc.2015.07.059 BindingDB Entry DOI: 10.7270/Q2HX1FF5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (232/232 = 100%)† (Escherichia coli (strain K12)) | BDBM22113 ((3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-y...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.0730 | -57.9 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Albert Einstein College of Medicine | Assay Description Purified MTAN activity in MTAN enzyme inhibition assay | Nat Chem Biol 5: 251-7 (2009) Article DOI: 10.1038/nchembio.153 BindingDB Entry DOI: 10.7270/Q28C9TM9 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (232/232 = 100%)† (Escherichia coli (strain K12)) | BDBM22113 ((3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-y...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 0.00200 | n/a | n/a | n/a | 7.5 | 25 |
Albert Einstein College of Medicine | Assay Description Enzyme activity assay using various bacterial species of 5'-Methylthioadenosine nucleosidases (MTANs) excepted for MTAP, for human MTAP. | ACS Chem Biol 2: 725-34 (2007) Article DOI: 10.1021/cb700166z BindingDB Entry DOI: 10.7270/Q2QF8R7W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (210/232 = 91%)† (Klebsiella pneumoniae) | BDBM22113 ((3R,4S)-1-({4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-y...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 0.784 | n/a | n/a | n/a | 7.5 | 25 |
Albert Einstein College of Medicine | Assay Description Enzyme activity assay using various bacterial species of 5'-Methylthioadenosine nucleosidases (MTANs) excepted for MTAP, for human MTAP. | ACS Chem Biol 2: 725-34 (2007) Article DOI: 10.1021/cb700166z BindingDB Entry DOI: 10.7270/Q2QF8R7W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||