

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-phosphoinositide-dependent protein kinase 1 (286/286 = 100%)† (Homo sapiens (Human)) | BDBM16996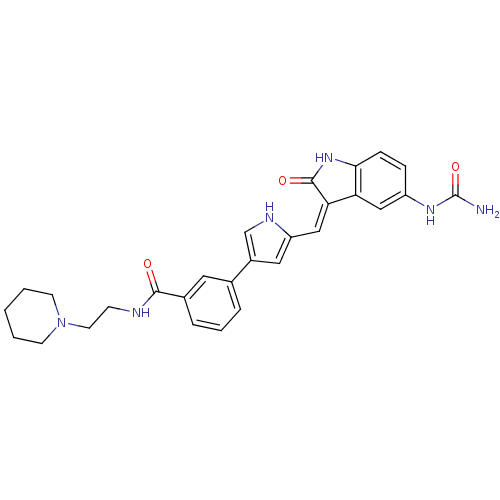 (3-(5-{[(3Z)-5-(carbamoylamino)-2-oxo-2,3-dihydro-1...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 4 | n/a | n/a | n/a | n/a | 7.2 | 22 |
Berlex Biosciences | Assay Description The coupled assay can detect inhibitors of AKT2 activation, as well as direct inhibitors of PDK1 or AKT2. Inactive AKT2 is activated in situ by incub... | Bioorg Med Chem Lett 17: 3819-25 (2007) Article DOI: 10.1016/j.bmcl.2007.05.060 BindingDB Entry DOI: 10.7270/Q29G5K22 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||