

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbonic anhydrase 2 (259/259 = 100%)† (Homo sapiens (Human)) | BDBM50200940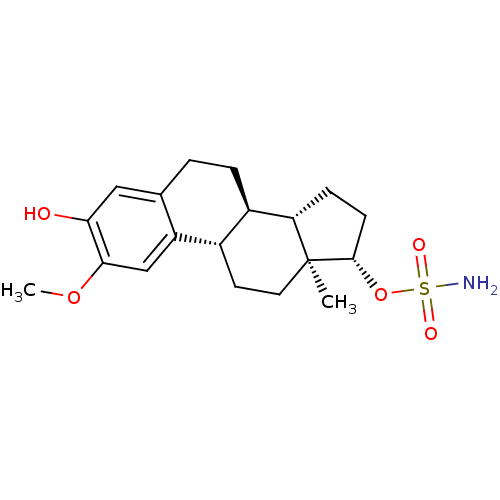 ((13ALPHA,14BETA,17ALPHA)-3-HYDROXY-2-METHOXYESTRA-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 526 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Bath Curated by ChEMBL | Assay Description Inhibition of human carbonic anhydrase 2 | J Med Chem 51: 1295-308 (2008) Article DOI: 10.1021/jm701319c BindingDB Entry DOI: 10.7270/Q2M0468S | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Carbonic anhydrase 2 (259/259 = 100%)† (Homo sapiens (Human)) | BDBM50200940 ((13ALPHA,14BETA,17ALPHA)-3-HYDROXY-2-METHOXYESTRA-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 526 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Bath Curated by ChEMBL | Assay Description Inhibition of human CA2 | J Med Chem 49: 7683-96 (2006) Article DOI: 10.1021/jm060705x BindingDB Entry DOI: 10.7270/Q2Z89C2X | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||