

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690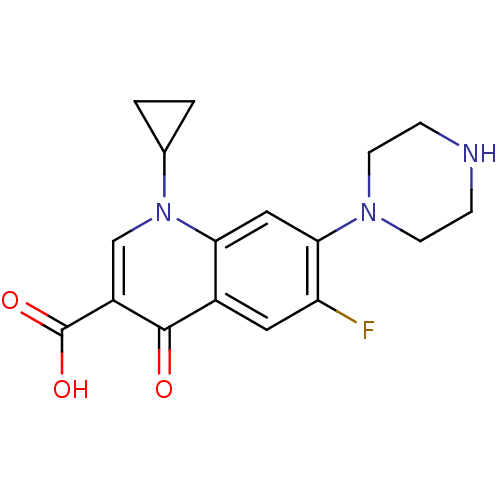 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 4.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase supercoiling activity using pBR322 as substrate after 30 mins by agarose gel electrophoresis | Bioorg Med Chem Lett 28: 2477-2480 (2018) Article DOI: 10.1016/j.bmcl.2018.06.003 BindingDB Entry DOI: 10.7270/Q2D2218S | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 1.05E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Medical University of Warsaw Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase supercoiling activity using pBR322 DNA as substrate after 30 mins by ethidium bromide staining based a... | Eur J Med Chem 156: 631-640 (2018) Article DOI: 10.1016/j.ejmech.2018.07.041 BindingDB Entry DOI: 10.7270/Q2VQ357Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 1.49E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Anhui Medical University Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase subunit A supercoiling activity using relaxed pBR322 DNA as substrate after 60 mins by agarose gel ele... | Eur J Med Chem 154: 144-154 (2018) Article DOI: 10.1016/j.ejmech.2018.05.021 BindingDB Entry DOI: 10.7270/Q2833VMP | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 3.15E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
University of Minnesota Medical School Curated by ChEMBL | Assay Description Inhibition of DNA supercoiling activity of Staphylococcus aureus DNA gyrase | Antimicrob Agents Chemother 54: 3011-4 (2010) Article DOI: 10.1128/AAC.00190-10 BindingDB Entry DOI: 10.7270/Q2W098SQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 3.92E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Daiichi Sankyo Co., Ltd Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus wild type DNA gyrase | Bioorg Med Chem 17: 6879-89 (2009) Article DOI: 10.1016/j.bmc.2009.08.026 BindingDB Entry DOI: 10.7270/Q2M90CHK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 6.17E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Achillion Pharmaceuticals Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus wild-type DNA gyrase supercoiling activity assessed as effect on conversion of relaxed pBR322 DNA to supercoiled ... | Antimicrob Agents Chemother 51: 2445-53 (2007) Article DOI: 10.1128/aac.00158-07 BindingDB Entry DOI: 10.7270/Q2QR50XQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 6.20E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Achillion Pharmaceuticals, Inc. Curated by ChEMBL | Assay Description Inhibition of wild-type Staphylococcus aureus DNA gyrase assessed as inhibition of supercoiling of pBR322 DNA after 60 min by gel electrophoresis ass... | J Med Chem 54: 3268-82 (2011) Article DOI: 10.1021/jm101604v BindingDB Entry DOI: 10.7270/Q28S4SRX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 6.20E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Achillion Pharmaceuticals, Inc. Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase by supercoiling assay | J Med Chem 50: 199-210 (2007) Article DOI: 10.1021/jm060844e BindingDB Entry DOI: 10.7270/Q2R78J0H | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 1.28E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Actelion Pharmaceuticals Ltd. Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus N-terminal His6-tagged DNA gyrase subunit GyrA/GyrB supercoiling activity expressed in Escherichia coli BL21 (DE3... | J Med Chem 60: 3755-3775 (2017) Article DOI: 10.1021/acs.jmedchem.6b01834 BindingDB Entry DOI: 10.7270/Q2SQ92VF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 1.28E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Actelion Pharmaceuticals Ltd Curated by ChEMBL | Assay Description Inhibition of wild type Staphylococcus aureus DNA gyrase subunit 2GyrA/2GyrB assessed as pBR322 supercoiling after 1 hr | J Med Chem 56: 7396-415 (2013) Article DOI: 10.1021/jm400963y BindingDB Entry DOI: 10.7270/Q2HT2QRV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Achillion Pharmaceuticals, Inc. Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase Ser84Leu mutant assessed as inhibition of supercoiling of pBR322 DNA after 60 min by gel electrophores... | J Med Chem 54: 3268-82 (2011) Article DOI: 10.1021/jm101604v BindingDB Entry DOI: 10.7270/Q28S4SRX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | >3.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Achillion Pharmaceuticals Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase gyrA S84L mutant supercoiling activity assessed as effect on conversion of relaxed pBR322 DNA to super... | Antimicrob Agents Chemother 51: 2445-53 (2007) Article DOI: 10.1128/aac.00158-07 BindingDB Entry DOI: 10.7270/Q2QR50XQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A (488/491 > 99%)† (Staphylococcus aureus) | BDBM21690 (1-cyclopropyl-6-fluoro-4-oxo-7-(piperazin-1-yl)-1,...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | >7.72E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Daiichi Sankyo Co., Ltd Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase Ser84Leu mutant | Bioorg Med Chem 17: 6879-89 (2009) Article DOI: 10.1016/j.bmc.2009.08.026 BindingDB Entry DOI: 10.7270/Q2M90CHK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||