 Found 6 hits Enzyme Inhibition Constant Data
Found 6 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Focal adhesion kinase 1
(276/276 = 100%)†
(Homo sapiens (Human)) | BDBM50318884
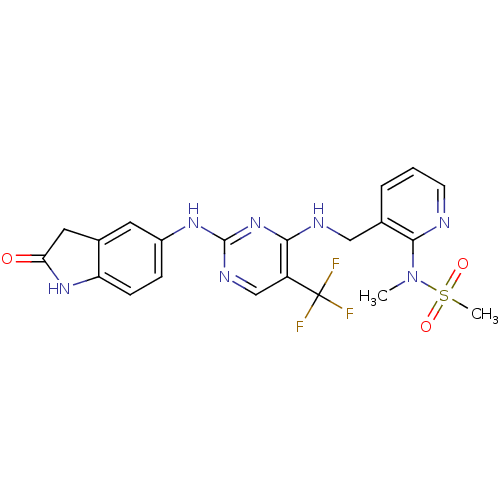
(CHEMBL1084546 | CHEMBL2430359 | N-methyl-N-(3-((2-...)Show SMILES CN(c1ncccc1CNc1nc(Nc2ccc3NC(=O)Cc3c2)ncc1C(F)(F)F)S(C)(=O)=O Show InChI InChI=1S/C21H20F3N7O3S/c1-31(35(2,33)34)19-12(4-3-7-25-19)10-26-18-15(21(22,23)24)11-27-20(30-18)28-14-5-6-16-13(8-14)9-17(32)29-16/h3-8,11H,9-10H2,1-2H3,(H,29,32)(H2,26,27,28,30) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 1 | n/a | n/a | n/a | n/a | n/a | n/a |
Lilly Research Laboratories
Curated by ChEMBL
| Assay Description
Inhibition of PTK2 (unknown origin) |
J Med Chem 56: 6991-7002 (2013)
Article DOI: 10.1021/jm400798j
BindingDB Entry DOI: 10.7270/Q2G44RQC |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Focal adhesion kinase 1
(276/276 = 100%)†
(Homo sapiens (Human)) | BDBM50318884

(CHEMBL1084546 | CHEMBL2430359 | N-methyl-N-(3-((2-...)Show SMILES CN(c1ncccc1CNc1nc(Nc2ccc3NC(=O)Cc3c2)ncc1C(F)(F)F)S(C)(=O)=O Show InChI InChI=1S/C21H20F3N7O3S/c1-31(35(2,33)34)19-12(4-3-7-25-19)10-26-18-15(21(22,23)24)11-27-20(30-18)28-14-5-6-16-13(8-14)9-17(32)29-16/h3-8,11H,9-10H2,1-2H3,(H,29,32)(H2,26,27,28,30) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 1.5 | n/a | n/a | n/a | n/a | n/a | n/a |
Punjabi University
Curated by ChEMBL
| Assay Description
Reversible inhibition of NH2-terminal 6His-tagged FAK catalytic domain (410 to 689 residues) (unknown origin) expressed in Sf9 insect cells using p(G... |
Eur J Med Chem 134: 159-184 (2017)
Article DOI: 10.1016/j.ejmech.2017.04.003
BindingDB Entry DOI: 10.7270/Q23X88SH |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Focal adhesion kinase 1
(276/276 = 100%)†
(Homo sapiens (Human)) | BDBM50318884

(CHEMBL1084546 | CHEMBL2430359 | N-methyl-N-(3-((2-...)Show SMILES CN(c1ncccc1CNc1nc(Nc2ccc3NC(=O)Cc3c2)ncc1C(F)(F)F)S(C)(=O)=O Show InChI InChI=1S/C21H20F3N7O3S/c1-31(35(2,33)34)19-12(4-3-7-25-19)10-26-18-15(21(22,23)24)11-27-20(30-18)28-14-5-6-16-13(8-14)9-17(32)29-16/h3-8,11H,9-10H2,1-2H3,(H,29,32)(H2,26,27,28,30) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 1.5 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck KGaA
Curated by ChEMBL
| Assay Description
Competitive binding affinity to FAK kinase domain (410 to 689) (unknown origin) assessed as phosphorylation of p(Glu/Tyr) in presence of ATP |
Bioorg Med Chem Lett 23: 5401-9 (2013)
Article DOI: 10.1016/j.bmcl.2013.07.050
BindingDB Entry DOI: 10.7270/Q2891782 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Focal adhesion kinase 1
(276/276 = 100%)†
(Homo sapiens (Human)) | BDBM50318884

(CHEMBL1084546 | CHEMBL2430359 | N-methyl-N-(3-((2-...)Show SMILES CN(c1ncccc1CNc1nc(Nc2ccc3NC(=O)Cc3c2)ncc1C(F)(F)F)S(C)(=O)=O Show InChI InChI=1S/C21H20F3N7O3S/c1-31(35(2,33)34)19-12(4-3-7-25-19)10-26-18-15(21(22,23)24)11-27-20(30-18)28-14-5-6-16-13(8-14)9-17(32)29-16/h3-8,11H,9-10H2,1-2H3,(H,29,32)(H2,26,27,28,30) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 1.5 | n/a | n/a | n/a | n/a | n/a | n/a |
Gilead Sciences, Inc.
Curated by ChEMBL
| Assay Description
Reversible/competitive inhibition of NH2-terminal His6-tagged FAK kinase domain (410 to 689 residues) (unknown origin) expressed in baculovirus infec... |
Bioorg Med Chem Lett 26: 5926-5930 (2016)
Article DOI: 10.1016/j.bmcl.2016.10.092
BindingDB Entry DOI: 10.7270/Q2ST7RTJ |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Focal adhesion kinase 1
(276/276 = 100%)†
(Homo sapiens (Human)) | BDBM50318884

(CHEMBL1084546 | CHEMBL2430359 | N-methyl-N-(3-((2-...)Show SMILES CN(c1ncccc1CNc1nc(Nc2ccc3NC(=O)Cc3c2)ncc1C(F)(F)F)S(C)(=O)=O Show InChI InChI=1S/C21H20F3N7O3S/c1-31(35(2,33)34)19-12(4-3-7-25-19)10-26-18-15(21(22,23)24)11-27-20(30-18)28-14-5-6-16-13(8-14)9-17(32)29-16/h3-8,11H,9-10H2,1-2H3,(H,29,32)(H2,26,27,28,30) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 1.5 | n/a | n/a | n/a | n/a | n/a | n/a |
Amgen, Inc.
Curated by ChEMBL
| Assay Description
Inhibition of FAK |
J Med Chem 53: 4332-53 (2010)
Article DOI: 10.1021/jm9018756
BindingDB Entry DOI: 10.7270/Q21R6QPF |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Focal adhesion kinase 1
(276/276 = 100%)†
(Homo sapiens (Human)) | BDBM50318884

(CHEMBL1084546 | CHEMBL2430359 | N-methyl-N-(3-((2-...)Show SMILES CN(c1ncccc1CNc1nc(Nc2ccc3NC(=O)Cc3c2)ncc1C(F)(F)F)S(C)(=O)=O Show InChI InChI=1S/C21H20F3N7O3S/c1-31(35(2,33)34)19-12(4-3-7-25-19)10-26-18-15(21(22,23)24)11-27-20(30-18)28-14-5-6-16-13(8-14)9-17(32)29-16/h3-8,11H,9-10H2,1-2H3,(H,29,32)(H2,26,27,28,30) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
| n/a | n/a | 1.5 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA
| Assay Description
Reversible inhibition of FAK kinase domain (amino acid 410-689) (unknown origin) assessed as inhibition of Phosphorylation of p(Glu/Tyr) by spectroph... |
Citation and Details
Article DOI: 10.1007/s00044-013-0768-0
BindingDB Entry DOI: 10.7270/Q2XW4M8H |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data