 Found 9 hits Enzyme Inhibition Constant Data
Found 9 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887
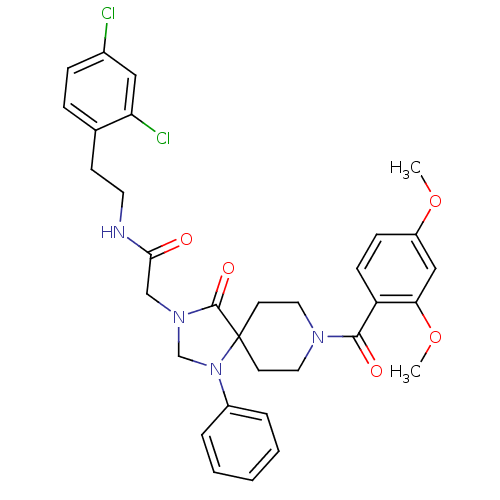
(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 650 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 900 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 1.50E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 2.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 2.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | 5.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | >5.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | >1.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
Dihydrolipoyl dehydrogenase
(464/464 = 100%)†
(Mycobacterium tuberculosis) | BDBM92887

(Triazaspirobenzyl, 5)Show SMILES COc1ccc(C(=O)N2CCC3(CC2)N(CN(CC(=O)NCCc2ccc(Cl)cc2Cl)C3=O)c2ccccc2)c(OC)c1 Show InChI InChI=1S/C32H34Cl2N4O5/c1-42-25-10-11-26(28(19-25)43-2)30(40)36-16-13-32(14-17-36)31(41)37(21-38(32)24-6-4-3-5-7-24)20-29(39)35-15-12-22-8-9-23(33)18-27(22)34/h3-11,18-19H,12-17,20-21H2,1-2H3,(H,35,39) | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| MMDB
PC cid
PC sid
PDB
UniChem
Similars
| MMDB
PDB
Article
PubMed
| n/a | n/a | >1.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Weill Cornell Medical College
| Assay Description
Enzymatic assay using Lpd. |
Biochemistry 49: 1616-27 (2010)
Article DOI: 10.1021/bi9016186
BindingDB Entry DOI: 10.7270/Q2JW8CGD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data