 Found 4 hits Enzyme Inhibition Constant Data
Found 4 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
E3 ubiquitin-protein ligase Mdm2
(93/94 = 99%)†
(Homo sapiens (Human)) | BDBM50436047
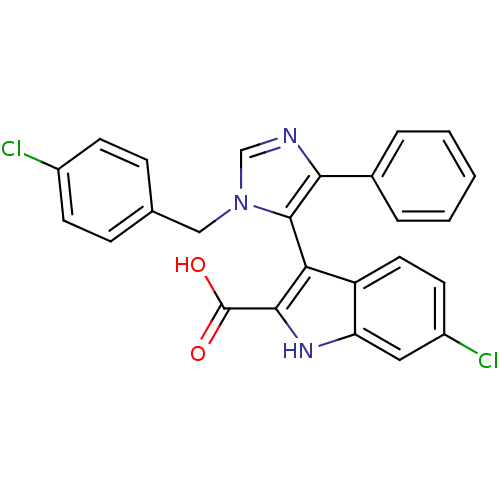
(CHEMBL1233798)Show SMILES OC(=O)c1[nH]c2cc(Cl)ccc2c1-c1c(ncn1Cc1ccc(Cl)cc1)-c1ccccc1 |(-2.71,1.13,;-1.94,2.47,;-2.71,3.8,;-.4,2.47,;.5,3.71,;1.96,3.24,;3.3,4.01,;4.63,3.24,;5.97,4.01,;4.63,1.7,;3.3,.93,;1.96,1.7,;.5,1.22,;.02,-.24,;.93,-1.49,;.02,-2.73,;-1.44,-2.26,;-1.44,-.72,;-2.69,.19,;-4.09,-.44,;-4.25,-1.97,;-5.66,-2.6,;-6.91,-1.69,;-8.31,-2.32,;-6.75,-.16,;-5.34,.47,;2.47,-1.49,;3.24,-.15,;4.78,-.15,;5.55,-1.49,;4.78,-2.82,;3.24,-2.82,)| Show InChI InChI=1S/C25H17Cl2N3O2/c26-17-8-6-15(7-9-17)13-30-14-28-22(16-4-2-1-3-5-16)24(30)21-19-11-10-18(27)12-20(19)29-23(21)25(31)32/h1-12,14,29H,13H2,(H,31,32) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| MMDB
PDB
Article
PubMed
| 916 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Utah
Curated by ChEMBL
| Assay Description
Displacement of fluorescent P4 peptide from recombinant human MDM2 (18 to 125) expressed in Escherichia coli BL21(DE3) RIL expression system after 30... |
Bioorg Med Chem Lett 24: 2546-54 (2014)
Article DOI: 10.1016/j.bmcl.2014.03.095
BindingDB Entry DOI: 10.7270/Q2FX7C05 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(93/94 = 99%)†
(Homo sapiens (Human)) | BDBM50436047

(CHEMBL1233798)Show SMILES OC(=O)c1[nH]c2cc(Cl)ccc2c1-c1c(ncn1Cc1ccc(Cl)cc1)-c1ccccc1 |(-2.71,1.13,;-1.94,2.47,;-2.71,3.8,;-.4,2.47,;.5,3.71,;1.96,3.24,;3.3,4.01,;4.63,3.24,;5.97,4.01,;4.63,1.7,;3.3,.93,;1.96,1.7,;.5,1.22,;.02,-.24,;.93,-1.49,;.02,-2.73,;-1.44,-2.26,;-1.44,-.72,;-2.69,.19,;-4.09,-.44,;-4.25,-1.97,;-5.66,-2.6,;-6.91,-1.69,;-8.31,-2.32,;-6.75,-.16,;-5.34,.47,;2.47,-1.49,;3.24,-.15,;4.78,-.15,;5.55,-1.49,;4.78,-2.82,;3.24,-2.82,)| Show InChI InChI=1S/C25H17Cl2N3O2/c26-17-8-6-15(7-9-17)13-30-14-28-22(16-4-2-1-3-5-16)24(30)21-19-11-10-18(27)12-20(19)29-23(21)25(31)32/h1-12,14,29H,13H2,(H,31,32) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| MMDB
PDB
Article
PubMed
| 916 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Michigan Comprehensive Cancer Center and Departments of Internal Medicine
Curated by ChEMBL
| Assay Description
Binding affinity to human MDM2 by fluorescence polarization assay |
J Med Chem 58: 1038-52 (2015)
Article DOI: 10.1021/jm501092z
BindingDB Entry DOI: 10.7270/Q21G0NZ5 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(93/94 = 99%)†
(Homo sapiens (Human)) | BDBM50436047

(CHEMBL1233798)Show SMILES OC(=O)c1[nH]c2cc(Cl)ccc2c1-c1c(ncn1Cc1ccc(Cl)cc1)-c1ccccc1 |(-2.71,1.13,;-1.94,2.47,;-2.71,3.8,;-.4,2.47,;.5,3.71,;1.96,3.24,;3.3,4.01,;4.63,3.24,;5.97,4.01,;4.63,1.7,;3.3,.93,;1.96,1.7,;.5,1.22,;.02,-.24,;.93,-1.49,;.02,-2.73,;-1.44,-2.26,;-1.44,-.72,;-2.69,.19,;-4.09,-.44,;-4.25,-1.97,;-5.66,-2.6,;-6.91,-1.69,;-8.31,-2.32,;-6.75,-.16,;-5.34,.47,;2.47,-1.49,;3.24,-.15,;4.78,-.15,;5.55,-1.49,;4.78,-2.82,;3.24,-2.82,)| Show InChI InChI=1S/C25H17Cl2N3O2/c26-17-8-6-15(7-9-17)13-30-14-28-22(16-4-2-1-3-5-16)24(30)21-19-11-10-18(27)12-20(19)29-23(21)25(31)32/h1-12,14,29H,13H2,(H,31,32) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| MMDB
PDB
Article
PubMed
| 920 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Pittsburgh
Curated by ChEMBL
| Assay Description
Binding affinity to MDM2 (unknown origin) expressed in Escherichia coli BL21(DE3) after 30 mins by fluorescence polarization assay in presence of P4 ... |
Bioorg Med Chem 21: 3982-95 (2013)
Article DOI: 10.1016/j.bmc.2012.06.020
BindingDB Entry DOI: 10.7270/Q2QV3NWD |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
E3 ubiquitin-protein ligase Mdm2
(93/94 = 99%)†
(Homo sapiens (Human)) | BDBM50436047

(CHEMBL1233798)Show SMILES OC(=O)c1[nH]c2cc(Cl)ccc2c1-c1c(ncn1Cc1ccc(Cl)cc1)-c1ccccc1 |(-2.71,1.13,;-1.94,2.47,;-2.71,3.8,;-.4,2.47,;.5,3.71,;1.96,3.24,;3.3,4.01,;4.63,3.24,;5.97,4.01,;4.63,1.7,;3.3,.93,;1.96,1.7,;.5,1.22,;.02,-.24,;.93,-1.49,;.02,-2.73,;-1.44,-2.26,;-1.44,-.72,;-2.69,.19,;-4.09,-.44,;-4.25,-1.97,;-5.66,-2.6,;-6.91,-1.69,;-8.31,-2.32,;-6.75,-.16,;-5.34,.47,;2.47,-1.49,;3.24,-.15,;4.78,-.15,;5.55,-1.49,;4.78,-2.82,;3.24,-2.82,)| Show InChI InChI=1S/C25H17Cl2N3O2/c26-17-8-6-15(7-9-17)13-30-14-28-22(16-4-2-1-3-5-16)24(30)21-19-11-10-18(27)12-20(19)29-23(21)25(31)32/h1-12,14,29H,13H2,(H,31,32) | PDB
MMDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MMDB
PC cid
PC sid
PDB
UniChem
| MMDB
PDB
Article
PubMed
| 920 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Groningen
Curated by ChEMBL
| Assay Description
Competition binding affinity to MDM2 (unknown origin) using p53 mimicking peptide TSFAEYWNLLSP after 30 mins by fluorescence polarization assay |
Bioorg Med Chem Lett 25: 5661-6 (2015)
Article DOI: 10.1016/j.bmcl.2015.11.019
BindingDB Entry DOI: 10.7270/Q2WM1HF9 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data